5WWM
 
 | |
5WYL
 
 | |
7V4Y
 
 | | TTHA1264/TTHA1265 complex | | 分子名称: | Putative zinc protease, ZINC ION, Zinc-dependent peptidase | | 著者 | Xu, M, Xu, Q, Ran, T, Wang, W, Sun, B, Wang, Q. | | 登録日 | 2021-08-16 | | 公開日 | 2022-06-29 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Crystal structures of TTHA1265 and TTHA1264/TTHA1265 complex reveal an intrinsic heterodimeric assembly.
Int.J.Biol.Macromol., 207, 2022
|
|
6M1J
 
 | | The DNA Gyrase B ATP binding domain of PSEUDOMONAS AERUGINOSA in complex with compound 12x | | 分子名称: | 1-[5-[6-fluoranyl-8-(methylamino)-4-[3-(trifluoromethyl)pyrazol-1-yl]-9H-pyrido[2,3-b]indol-3-yl]pyrimidin-2-yl]cyclopropane-1-carboxylic acid, DIMETHYL SULFOXIDE, DNA gyrase subunit B, ... | | 著者 | Xu, Z.H, Zhou, Z. | | 登録日 | 2020-02-26 | | 公開日 | 2020-09-02 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.701 Å) | | 主引用文献 | Discovery of Pyrido[2,3-b]indole Derivatives with Gram-Negative Activity Targeting Both DNA Gyrase and Topoisomerase IV.
J.Med.Chem., 63, 2020
|
|
6B70
 
 | | Cryo-EM structure of human insulin degrading enzyme in complex with FAB H11-E heavy chain, FAB H11-E light chain and insulin | | 分子名称: | FAB H11-E heavy chain, FAB H11-E light chain, Insulin, ... | | 著者 | Liang, W.G, Zhang, Z, Bailey, L.J, Kossiakoff, A.A, Tan, Y.Z, Wei, H, Carragher, B, Potter, S.C, Tang, W.J. | | 登録日 | 2017-10-03 | | 公開日 | 2017-12-27 | | 最終更新日 | 2021-04-28 | | 実験手法 | ELECTRON MICROSCOPY (3.7 Å) | | 主引用文献 | Ensemble cryoEM elucidates the mechanism of insulin capture and degradation by human insulin degrading enzyme.
Elife, 7, 2018
|
|
5B77
 
 | |
5B78
 
 | |
5ZJI
 
 | | Structure of photosystem I supercomplex with light-harvesting complexes I and II | | 分子名称: | (1R,3R)-6-{(3E,5E,7E,9E,11E,13E,15E,17E)-18-[(1S,4R,6R)-4-HYDROXY-2,2,6-TRIMETHYL-7-OXABICYCLO[4.1.0]HEPT-1-YL]-3,7,12,16-TETRAMETHYLOCTADECA-1,3,5,7,9,11,13,15,17-NONAENYLIDENE}-1,5,5-TRIMETHYLCYCLOHEXANE-1,3-DIOL, (3R,3'R,6S)-4,5-DIDEHYDRO-5,6-DIHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, (3S,5R,6S,3'S,5'R,6'S)-5,6,5',6'-DIEPOXY-5,6,5',6'- TETRAHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, ... | | 著者 | Pan, X.W, Ma, J, Su, X.D, Cao, P, Liu, Z.F, Zhang, X.Z, Li, M. | | 登録日 | 2018-03-20 | | 公開日 | 2018-06-20 | | 最終更新日 | 2019-11-06 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | Structure of the maize photosystem I supercomplex with light-harvesting complexes I and II.
Science, 360, 2018
|
|
5IE3
 
 | | Crystal structure of a plant enzyme | | 分子名称: | ADENOSINE MONOPHOSPHATE, OXALIC ACID, Oxalate--CoA ligase | | 著者 | Fan, M.R, Li, M, Chang, W.R. | | 登録日 | 2016-02-24 | | 公開日 | 2016-12-07 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Crystal Structures of Arabidopsis thaliana Oxalyl-CoA Synthetase Essential for Oxalate Degradation
Mol Plant, 9, 2016
|
|
5B79
 
 | | Crystal structure of DPF2 double PHD finger | | 分子名称: | ZINC ION, Zinc finger protein ubi-d4 | | 著者 | Li, H, Xiong, X. | | 登録日 | 2016-06-05 | | 公開日 | 2016-10-26 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.601 Å) | | 主引用文献 | Selective recognition of histone crotonylation by double PHD fingers of MOZ and DPF2
Nat.Chem.Biol., 12, 2016
|
|
7YV8
 
 | | Cryo-EM structure of SARS-CoV-2 Omicron BA.2 RBD in complex with golden hamster ACE2 (local refinement) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme, Spike glycoprotein, ... | | 著者 | Zhao, Z.N, Xie, Y.F, Chai, Y, Qi, J.X, Gao, G.F. | | 登録日 | 2022-08-18 | | 公開日 | 2023-07-19 | | 最終更新日 | 2023-08-02 | | 実験手法 | ELECTRON MICROSCOPY (2.94 Å) | | 主引用文献 | Structural basis for receptor binding and broader interspecies receptor recognition of currently circulating Omicron sub-variants.
Nat Commun, 14, 2023
|
|
7YVU
 
 | | Cryo-EM structure of SARS-CoV-2 Omicron BA.2 RBD in complex with mouse ACE2 (local refinement) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, Spike protein S1, ... | | 著者 | Zhao, Z.N, Xie, Y.F, Chai, Y, Qi, J.X, Gao, G.F. | | 登録日 | 2022-08-19 | | 公開日 | 2023-07-19 | | 最終更新日 | 2023-08-02 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Structural basis for receptor binding and broader interspecies receptor recognition of currently circulating Omicron sub-variants.
Nat Commun, 14, 2023
|
|
1XP6
 
 | | HUMAN ESTROGEN RECEPTOR ALPHA LIGAND-BINDING DOMAIN IN COMPLEX WITH COMPOUND 16 | | 分子名称: | (2S,3R)-2-(4-{2-[(3S,4S)-3,4-DIMETHYLPYRROLIDIN-1-YL]ETHOXY}PHENYL)-3-(4-HYDROXYPHENYL)-2,3-DIHYDRO-1,4-BENZOXATHIIN-6-OL, Estrogen receptor | | 著者 | Fitzgerald, P.M.D, Sharma, N. | | 登録日 | 2004-10-08 | | 公開日 | 2004-12-07 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Estrogen receptor ligands. Part 9: Dihydrobenzoxathiin SERAMs with alkyl substituted pyrrolidine side chains and linkers.
Bioorg.Med.Chem.Lett., 15, 2005
|
|
5O6T
 
 | | BIRC4 RING in complex with dimeric ubiquitin variant | | 分子名称: | 1,2-ETHANEDIOL, E3 ubiquitin-protein ligase XIAP, Polyubiquitin-B, ... | | 著者 | Gabrielsen, M, Buetow, L, Huang, D.T. | | 登録日 | 2017-06-07 | | 公開日 | 2017-11-01 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.57 Å) | | 主引用文献 | A General Strategy for Discovery of Inhibitors and Activators of RING and U-box E3 Ligases with Ubiquitin Variants.
Mol. Cell, 68, 2017
|
|
6B7Y
 
 | | Cryo-EM structure of human insulin degrading enzyme | | 分子名称: | Insulin-degrading enzyme | | 著者 | Liang, W.G, Zhang, Z, Bailey, L.J, Kossiakoff, A.A, Tan, Y.Z, Wei, H, Carragher, B, Potter, S.C, Tang, W.J. | | 登録日 | 2017-10-05 | | 公開日 | 2017-11-08 | | 最終更新日 | 2024-03-13 | | 実験手法 | ELECTRON MICROSCOPY (6.5 Å) | | 主引用文献 | Ensemble cryoEM elucidates the mechanism of insulin capture and degradation by human insulin degrading enzyme.
Elife, 7, 2018
|
|
6BFC
 
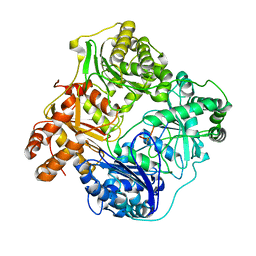 | | Cryo-EM structure of human insulin degrading enzyme in complex with insulin | | 分子名称: | Insulin, Insulin-degrading enzyme | | 著者 | Liang, W.G, Zhang, Z, Bailey, L.J, Kossiakoff, A.A, Tan, Y.Z, Wei, H, Carragher, B, Potter, S.C, Tang, W.J. | | 登録日 | 2017-10-26 | | 公開日 | 2017-12-27 | | 最終更新日 | 2021-04-28 | | 実験手法 | ELECTRON MICROSCOPY (3.7 Å) | | 主引用文献 | Ensemble cryoEM elucidates the mechanism of insulin capture and degradation by human insulin degrading enzyme.
Elife, 7, 2018
|
|
5YSW
 
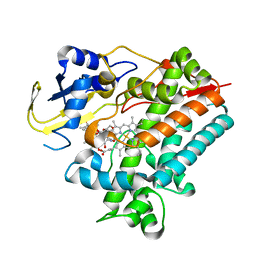 | | Crystal Structure Analysis of Rif16 in complex with R-L | | 分子名称: | (2S,12E,14E,16S,17S,18R,19R,20R,21S,22R,23S,24E)-21-(acetyloxy)-5,6,17,19-tetrahydroxy-23-methoxy-2,4,12,16,18,20,22-heptamethyl-1,11-dioxo-1,2-dihydro-2,7-(epoxypentadeca[1,11,13]trienoimino)naphtho[2,1-b]furan-9-yl hydroxyacetate, Cytochrome P450, PROTOPORPHYRIN IX CONTAINING FE | | 著者 | Li, F.W, Qi, F.F, Xiao, Y.L, Zhao, G.P, Li, S.Y. | | 登録日 | 2017-11-15 | | 公開日 | 2018-07-04 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Deciphering the late steps of rifamycin biosynthesis.
Nat Commun, 9, 2018
|
|
5O6S
 
 | |
5O75
 
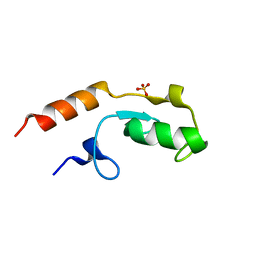 | | Ube4B U-box domain | | 分子名称: | SULFATE ION, Ubiquitin conjugation factor E4 B | | 著者 | Gabrielsen, M, Buetow, L, Nakasone, M.A, Huang, D.T. | | 登録日 | 2017-06-08 | | 公開日 | 2017-11-01 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.483 Å) | | 主引用文献 | A General Strategy for Discovery of Inhibitors and Activators of RING and U-box E3 Ligases with Ubiquitin Variants.
Mol. Cell, 68, 2017
|
|
5B75
 
 | |
6NSY
 
 | | X-ray reduced Catalase 3 From N.Crassa in Cpd I state (0.263 MGy) | | 分子名称: | 1,2-ETHANEDIOL, ACETATE ION, Catalase-3, ... | | 著者 | Zarate-Romero, A, Rudino-Pinera, E, Stojanoff, V. | | 登録日 | 2019-01-27 | | 公開日 | 2019-05-01 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | X-ray driven reduction of Cpd I of Catalase-3 from N. crassa reveals differential sensitivity of active sites and formation of ferrous state.
Arch.Biochem.Biophys., 666, 2019
|
|
6NX9
 
 | |
6NSZ
 
 | | X-ray reduced Catalase 3 from N.Crassa (0.526 MGy) | | 分子名称: | 1,2-ETHANEDIOL, ACETATE ION, Catalase-3, ... | | 著者 | Zarate-Romero, A, Rudino-Pinera, E, Stojanoff, V. | | 登録日 | 2019-01-27 | | 公開日 | 2019-05-01 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | X-ray driven reduction of Cpd I of Catalase-3 from N. crassa reveals differential sensitivity of active sites and formation of ferrous state.
Arch.Biochem.Biophys., 666, 2019
|
|
4ZLK
 
 | | Crystal structure of mouse myosin-5a in complex with calcium-bound calmodulin | | 分子名称: | CALCIUM ION, Calmodulin, Unconventional myosin-Va | | 著者 | Shen, M, Zhang, N, Zheng, S, Zhang, W.-B, Zhang, H.-M, Lu, Z, Su, Q.P, Sun, Y, Ye, K, Li, X.-D. | | 登録日 | 2015-05-01 | | 公開日 | 2016-05-04 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.502 Å) | | 主引用文献 | Structural basis for calcium regulation of myosin 5 motor function
To Be Published
|
|
6NT1
 
 | | Catalase 3 from N.Crassa in ferrous state (2.89 MGy) | | 分子名称: | 1,2-ETHANEDIOL, ACETATE ION, Catalase-3, ... | | 著者 | Zarate-Romero, A, Rudino-Pinera, E, Stojanoff, V. | | 登録日 | 2019-01-27 | | 公開日 | 2019-05-01 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | X-ray driven reduction of Cpd I of Catalase-3 from N. crassa reveals differential sensitivity of active sites and formation of ferrous state.
Arch.Biochem.Biophys., 666, 2019
|
|
