8XFQ
 
 | |
6PTX
 
 | | Dark, 100K, PCM Myxobacterial Phytochrome, P2, Wild Type, | | 分子名称: | 3-[(2Z)-2-({3-(2-carboxyethyl)-5-[(E)-(4-ethenyl-3-methyl-5-oxo-1,5-dihydro-2H-pyrrol-2-ylidene)methyl]-4-methyl-1H-pyrrol-2-yl}methylidene)-5-{(Z)-[(3E,4S)-3-ethylidene-4-methyl-5-oxopyrrolidin-2-ylidene]methyl}-4-methyl-2H-pyrrol-3-yl]propanoic acid, Photoreceptor-histidine kinase BphP | | 著者 | Pandey, S, Schmidt, M, Stojkovic, E.A. | | 登録日 | 2019-07-16 | | 公開日 | 2019-10-09 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | High-resolution crystal structures of a myxobacterial phytochrome at cryo and room temperatures.
Struct Dyn., 6, 2019
|
|
6WOQ
 
 | | Structure of Hepatitis C Virus Envelope Glycoprotein E2 core from genotype 1a bound to neutralizing antibody HC1AM and non neutralizing antibody E1 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein E2, ... | | 著者 | Tzarum, N, Wilson, I.A, Law, M. | | 登録日 | 2020-04-25 | | 公開日 | 2020-08-19 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (3.667 Å) | | 主引用文献 | An alternate conformation of HCV E2 neutralizing face as an additional vaccine target.
Sci Adv, 6, 2020
|
|
7URQ
 
 | |
7URS
 
 | |
6WO5
 
 | | Structure of Hepatitis C Virus Envelope Glycoprotein E2 core from genotype 1a bound to neutralizing antibody 212.1.1 and non neutralizing antibody E1 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein E2, ... | | 著者 | Tzarum, N, Wilson, I.A, Law, M. | | 登録日 | 2020-04-24 | | 公開日 | 2020-08-19 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.619 Å) | | 主引用文献 | An alternate conformation of HCV E2 neutralizing face as an additional vaccine target.
Sci Adv, 6, 2020
|
|
5LBR
 
 | | Wild-type PAS-GAF fragment from Deinococcus radiodurans Bphp collected at SACLA | | 分子名称: | 3-[2-[(Z)-[3-(2-carboxyethyl)-5-[(Z)-(4-ethenyl-3-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-4-methyl-pyrrol-1-ium -2-ylidene]methyl]-5-[(Z)-[(3E)-3-ethylidene-4-methyl-5-oxidanylidene-pyrrolidin-2-ylidene]methyl]-4-methyl-1H-pyrrol-3- yl]propanoic acid, ACETATE ION, Bacteriophytochrome | | 著者 | Edlund, P, Claesson, E, Nakane, T, Takala, H, Dods, R, Schmidt, M, Westenhoff, S. | | 登録日 | 2016-06-17 | | 公開日 | 2016-10-26 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | The room temperature crystal structure of a bacterial phytochrome determined by serial femtosecond crystallography.
Sci Rep, 6, 2016
|
|
6WO3
 
 | | Structure of Hepatitis C Virus Envelope Glycoprotein E2 core from genotype 6a bound to broadly neutralizing antibody U1 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein E2, ... | | 著者 | Tzarum, N, Wilson, I.A, Law, M. | | 登録日 | 2020-04-24 | | 公開日 | 2020-08-19 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.382 Å) | | 主引用文献 | An alternate conformation of HCV E2 neutralizing face as an additional vaccine target.
Sci Adv, 6, 2020
|
|
6WO4
 
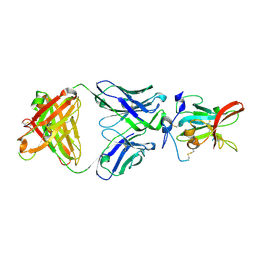 | | Structure of Hepatitis C Virus Envelope Glycoprotein E2 core from genotype 6a bound to broadly neutralizing antibody HC11 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein E2, ... | | 著者 | Tzarum, N, Wilson, I.A, Law, M. | | 登録日 | 2020-04-24 | | 公開日 | 2020-08-19 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.349 Å) | | 主引用文献 | An alternate conformation of HCV E2 neutralizing face as an additional vaccine target.
Sci Adv, 6, 2020
|
|
6Q2A
 
 | |
6WOR
 
 | |
6WOS
 
 | |
5L8M
 
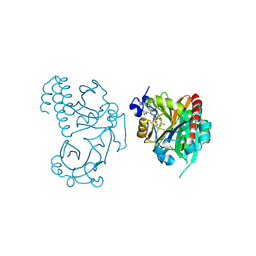 | | Wild-type PAS-GAF fragment from Deinococcus radiodurans Bphp collected at LCLS | | 分子名称: | 3-[2-[(Z)-[3-(2-carboxyethyl)-5-[(Z)-(4-ethenyl-3-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-4-methyl-pyrrol-1-ium -2-ylidene]methyl]-5-[(Z)-[(3E)-3-ethylidene-4-methyl-5-oxidanylidene-pyrrolidin-2-ylidene]methyl]-4-methyl-1H-pyrrol-3- yl]propanoic acid, ACETATE ION, Bacteriophytochrome | | 著者 | Claesson, E, Takala, H, Edlund, P, Henry, L, Dods, R, Schmidt, M, Westenhoff, S. | | 登録日 | 2016-06-08 | | 公開日 | 2016-10-26 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | The room temperature crystal structure of a bacterial phytochrome determined by serial femtosecond crystallography.
Sci Rep, 6, 2016
|
|
3QM0
 
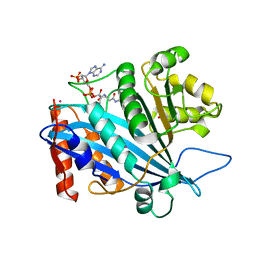 | | Crystal structure of RTT109-AC-CoA complex | | 分子名称: | ACETYL COENZYME *A, Histone acetyltransferase RTT109, MERCURY (II) ION | | 著者 | Tang, Y, Marmorstein, R. | | 登録日 | 2011-02-03 | | 公開日 | 2011-02-16 | | 最終更新日 | 2017-08-02 | | 実験手法 | X-RAY DIFFRACTION (3.1 Å) | | 主引用文献 | Fungal Rtt109 histone acetyltransferase is an unexpected structural homolog of metazoan p300/CBP.
Nat.Struct.Mol.Biol., 15, 2008
|
|
5EK0
 
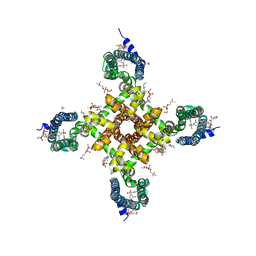 | | Human Nav1.7-VSD4-NavAb in complex with GX-936. | | 分子名称: | 1,2-DIMYRISTOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 3-cyano-4-[2-[2-(1-ethylazetidin-3-yl)pyrazol-3-yl]-4-(trifluoromethyl)phenoxy]-~{N}-(1,2,4-thiadiazol-5-yl)benzenesulfonamide, Chimera of bacterial Ion transport protein and human Sodium channel protein type 9 subunit alpha | | 著者 | Ahuja, S, Mukund, S, Starovasnik, M.A, Koth, C.M, Payandeh, J. | | 登録日 | 2015-11-03 | | 公開日 | 2015-12-23 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (3.53 Å) | | 主引用文献 | Structural basis of Nav1.7 inhibition by an isoform-selective small-molecule antagonist.
Science, 350, 2015
|
|
8A83
 
 | |
7QLL
 
 | |
7QLI
 
 | |
7QLK
 
 | |
6Q9Z
 
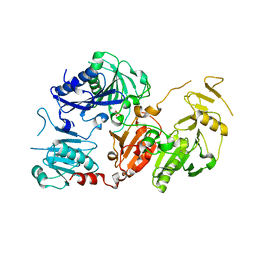 | | Crystal structure of the pathological G167R variant of calcium-free human gelsolin, | | 分子名称: | GLYCEROL, Gelsolin, SULFATE ION | | 著者 | Boni, F, Scalone, E, Milani, M, Eloise, M, de Rosa, M. | | 登録日 | 2018-12-18 | | 公開日 | 2019-11-27 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (3.8 Å) | | 主引用文献 | The structure of N184K amyloidogenic variant of gelsolin highlights the role of the H-bond network for protein stability and aggregation properties.
Eur.Biophys.J., 49, 2020
|
|
6QBF
 
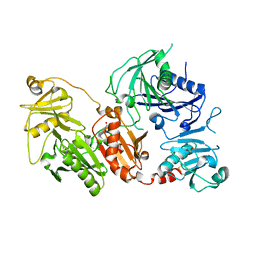 | | Crystal structure of the pathological D187N variant of calcium-free human gelsolin. | | 分子名称: | GLYCEROL, Gelsolin, SODIUM ION, ... | | 著者 | Scalone, E, Boni, F, Milani, M, Eloise, M, de Rosa, M. | | 登録日 | 2018-12-21 | | 公開日 | 2019-11-27 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (3.499 Å) | | 主引用文献 | The structure of N184K amyloidogenic variant of gelsolin highlights the role of the H-bond network for protein stability and aggregation properties.
Eur.Biophys.J., 49, 2020
|
|
6Q9R
 
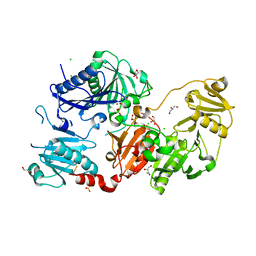 | | Crystal structure of the pathological N184K variant of calcium-free human gelsolin | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, GLYCEROL, ... | | 著者 | Scalone, E, Boni, F, Milani, M, Eloise, M, de Rosa, M. | | 登録日 | 2018-12-18 | | 公開日 | 2019-11-27 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.73 Å) | | 主引用文献 | The structure of N184K amyloidogenic variant of gelsolin highlights the role of the H-bond network for protein stability and aggregation properties.
Eur.Biophys.J., 49, 2020
|
|
8K6Y
 
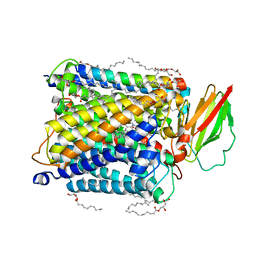 | | Serial femtosecond crystallography structure of photo dissociated CO from ba3- type cytochrome c oxidase determined by extrapolation method | | 分子名称: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, CARBON MONOXIDE, COPPER (II) ION, ... | | 著者 | Safari, C, Ghosh, S, Andersson, R, Johannesson, J, Donoso, A.V, Zoric, D, Sandelin, E, Iwata, S, Neutze, R, Branden, G. | | 登録日 | 2023-07-25 | | 公開日 | 2023-11-15 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Time-resolved serial crystallography to track the dynamics of carbon monoxide in the active site of cytochrome c oxidase.
Sci Adv, 9, 2023
|
|
7QLJ
 
 | |
7QLM
 
 | |
