3N9L
 
 | | ceKDM7A from C.elegans, complex with H3K4me3 peptide and NOG | | 分子名称: | FE (II) ION, Histone H3 peptide, N-OXALYLGLYCINE, ... | | 著者 | Yang, Y, Hu, L, Wang, P, Hou, H, Chen, C.D, Xu, Y. | | 登録日 | 2010-05-31 | | 公開日 | 2010-06-30 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.796 Å) | | 主引用文献 | Structural insights into a dual-specificity histone demethylase ceKDM7A from Caenorhabditis elegans
Cell Res., 20, 2010
|
|
5GZO
 
 | | Structure of neutralizing antibody bound to Zika envelope protein | | 分子名称: | Antibody heavy chain, Antibody light chain, Genome polyprotein | | 著者 | Wang, Q, Yang, H, Liu, X, Dai, L, Ma, T, Qi, J, Wong, G, Peng, R, Liu, S, Li, J, Li, S, Song, J, Liu, J, He, J, Yuan, H, Xiong, Y, Liao, Y, Li, J, Yang, J, Tong, Z, Griffin, B, Bi, Y, Liang, M, Xu, X, Cheng, G, Wang, P, Qiu, X, Kobinger, G, Shi, Y, Yan, J, Gao, G.F. | | 登録日 | 2016-09-29 | | 公開日 | 2017-01-18 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (2.755 Å) | | 主引用文献 | Molecular determinants of human neutralizing antibodies isolated from a patient infected with Zika virus
Sci Transl Med, 8, 2016
|
|
5GZN
 
 | | Structure of neutralizing antibody bound to Zika envelope protein | | 分子名称: | Antibody Heavy chain, Antibody light chain, Genome polyprotein | | 著者 | Wang, Q, Yang, H, Liu, X, Dai, L, Ma, T, Qi, J, Wong, G, Peng, R, Liu, S, Li, J, Li, S, Song, J, Liu, J, He, J, Yuan, H, Xiong, Y, Liao, Y, Li, J, Yang, J, Tong, Z, Griffin, B, Bi, Y, Liang, M, Xu, X, Cheng, G, Wang, P, Qiu, X, Kobinger, G, Shi, Y, Yan, J, Gao, G.F. | | 登録日 | 2016-09-29 | | 公開日 | 2016-12-28 | | 最終更新日 | 2024-10-23 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Molecular determinants of human neutralizing antibodies isolated from a patient infected with Zika virus
Sci Transl Med, 8, 2016
|
|
6IXL
 
 | |
6IXN
 
 | | Crystal structure of isocitrate dehydrogenase from Ostreococcus tauri in complex with NAD+ and citrate | | 分子名称: | CITRATE ANION, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | 著者 | Zhu, G.P, Tang, W.G, Wang, P. | | 登録日 | 2018-12-11 | | 公開日 | 2019-12-18 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (1.87 Å) | | 主引用文献 | Crystal structures of NAD + -linked isocitrate dehydrogenase from the green alga Ostreococcus tauri and its evolutionary relationship with eukaryotic NADP + -linked homologs.
Arch.Biochem.Biophys., 708, 2021
|
|
6IZS
 
 | | Crystal structure of Haemophilus influenzae BamA POTRA4 | | 分子名称: | Outer membrane protein assembly factor BamA | | 著者 | Ma, X, Wang, Q, Li, Y, Tan, P, Wu, H, Wang, P, Dong, X, Hong, L, Meng, G. | | 登録日 | 2018-12-20 | | 公開日 | 2019-10-30 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.03 Å) | | 主引用文献 | How BamA recruits OMP substratesviapoly-POTRAs domain.
Faseb J., 33, 2019
|
|
6IZT
 
 | | Crystal structure of Haemophilus Influenzae BamA POTRA3-5 | | 分子名称: | Outer membrane protein assembly factor BamA | | 著者 | Ma, X, Wang, Q, Li, Y, Tan, P, Wu, H, Wang, P, Dong, X, Hong, L, Meng, G. | | 登録日 | 2018-12-20 | | 公開日 | 2019-10-30 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.03 Å) | | 主引用文献 | How BamA recruits OMP substratesviapoly-POTRAs domain.
Faseb J., 33, 2019
|
|
5IAY
 
 | | NMR structure of UHRF1 Tandem Tudor Domains in a complex with Spacer peptide | | 分子名称: | E3 ubiquitin-protein ligase UHRF1, Spacer | | 著者 | Fang, J, Cheng, J, Wang, J, Zhang, Q, Liu, M, Gong, R, Wang, P, Zhang, X, Feng, Y, Lan, W, Gong, Z, Tang, C, Wong, J, Yang, H, Cao, C, Xu, Y. | | 登録日 | 2016-02-22 | | 公開日 | 2016-04-20 | | 最終更新日 | 2024-05-01 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Hemi-methylated DNA opens a closed conformation of UHRF1 to facilitate its histone recognition
Nat Commun, 7, 2016
|
|
6J09
 
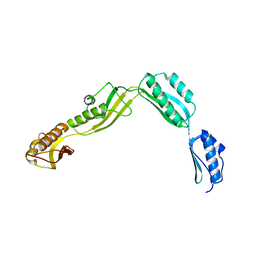 | | Crystal structure of Haemophilus Influenzae BamA POTRA1-4 | | 分子名称: | Outer membrane protein assembly factor BamA | | 著者 | Ma, X, Wang, Q, Li, Y, Tan, P, Wu, H, Wang, P, Dong, X, Hong, L, Meng, G. | | 登録日 | 2018-12-21 | | 公開日 | 2019-10-30 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | How BamA recruits OMP substratesviapoly-POTRAs domain.
Faseb J., 33, 2019
|
|
6IXT
 
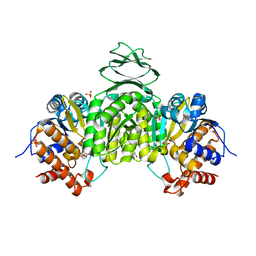 | | Crystal structure of isocitrate dehydrogenase from Ostreococcus tauri in complex with NAD+ and Mg2+ | | 分子名称: | GLYCEROL, Isocitrate dehydrogenase, MAGNESIUM ION, ... | | 著者 | Zhu, G.P, Tang, W.G, Wang, P. | | 登録日 | 2018-12-12 | | 公開日 | 2019-12-18 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (1.78 Å) | | 主引用文献 | Crystal structures of NAD + -linked isocitrate dehydrogenase from the green alga Ostreococcus tauri and its evolutionary relationship with eukaryotic NADP + -linked homologs.
Arch.Biochem.Biophys., 708, 2021
|
|
3R7W
 
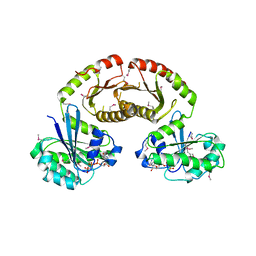 | | Crystal Structure of Gtr1p-Gtr2p complex | | 分子名称: | GTP-binding protein GTR1, GTP-binding protein GTR2, MAGNESIUM ION, ... | | 著者 | Gong, R, Li, L, Liu, Y, Wang, P, Yang, H, Wang, L, Cheng, J, Guan, K.L, Xu, Y. | | 登録日 | 2011-03-23 | | 公開日 | 2011-08-24 | | 最終更新日 | 2024-10-30 | | 実験手法 | X-RAY DIFFRACTION (2.773 Å) | | 主引用文献 | Crystal structure of the Gtr1p-Gtr2p complex reveals new insights into the amino acid-induced TORC1 activation
Genes Dev., 25, 2011
|
|
1NHX
 
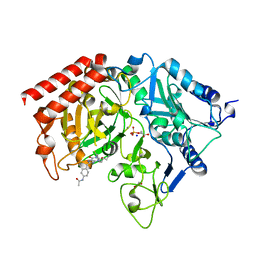 | | PEPCK COMPLEX WITH A GTP-COMPETITIVE INHIBITOR | | 分子名称: | 1,2-ETHANEDIOL, MANGANESE (II) ION, N-{4-[1-(2-FLUOROBENZYL)-3-BUTYL-2,6-DIOXO-2,3,6,7-TETRAHYDRO-1H-PURIN-8-YLMETHYL]-PHENYL}-ACETAMIDE, ... | | 著者 | Foley, L.H, Wang, P, Dunten, P, Ramsey, G, Gubler, M.-L, Wertheimer, S.J. | | 登録日 | 2002-12-19 | | 公開日 | 2003-09-30 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | X-RAY STRUCTURES OF TWO XANTHINE INHIBITORS BOUND TO PEPCK and N-3 modifications of substituted 1,8-Dibenzylxanthines
Bioorg.Med.Chem.Lett., 13, 2003
|
|
9KPN
 
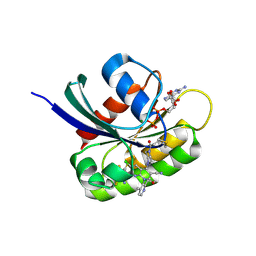 | | Crystal structure of KRAS-G12C in complex with Compound 20 (JAB-20) | | 分子名称: | 7-[2-azanyl-3,4,5,6-tetrakis(fluoranyl)phenyl]-6-chloranyl-1-(4-methyl-2-propan-2-yl-pyridin-3-yl)-2-oxidanylidene-4-(4-prop-2-enoylpiperazin-1-yl)-1,8-naphthyridine-3-carbonitrile, CHLORIDE ION, GUANOSINE-5'-DIPHOSPHATE, ... | | 著者 | Li, S, Dang, C, Fan, X, Zhang, W, Wang, P, Qian, D, Wang, Y, Li, A. | | 登録日 | 2024-11-23 | | 公開日 | 2025-03-12 | | 実験手法 | X-RAY DIFFRACTION (1.29 Å) | | 主引用文献 | Design, Structure Optimization, and Preclinical Characterization of JAB-21822, a Covalent Inhibitor of KRAS G12C.
J.Med.Chem., 68, 2025
|
|
9KPM
 
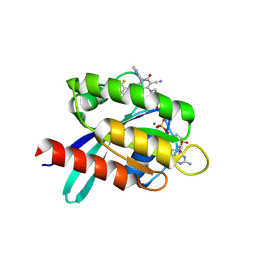 | | Crystal structure of KRAS-G12C in complex with compound 16 (JAB-16) | | 分子名称: | 7-[2-azanyl-3,5-bis(chloranyl)-6-fluoranyl-phenyl]-6-chloranyl-1-(4-methyl-2-propan-2-yl-pyridin-3-yl)-2-oxidanylidene-4-(4-prop-2-enoylpiperazin-1-yl)-1,8-naphthyridine-3-carbonitrile, GUANOSINE-5'-DIPHOSPHATE, Isoform 2B of GTPase KRas, ... | | 著者 | Li, S, Dang, C, Fan, X, Zhang, W, Wang, P, Qian, D, Wang, Y, Li, A. | | 登録日 | 2024-11-23 | | 公開日 | 2025-03-12 | | 実験手法 | X-RAY DIFFRACTION (1.41 Å) | | 主引用文献 | Design, Structure Optimization, and Preclinical Characterization of JAB-21822, a Covalent Inhibitor of KRAS G12C.
J.Med.Chem., 68, 2025
|
|
1M51
 
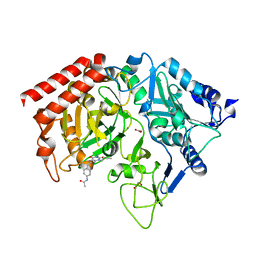 | | PEPCK complex with a GTP-competitive inhibitor | | 分子名称: | 1,2-ETHANEDIOL, ACETATE ION, MANGANESE (II) ION, ... | | 著者 | Foley, L.H, Wang, P, Dunten, P, Wertheimer, S.J. | | 登録日 | 2002-07-06 | | 公開日 | 2003-09-30 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2.25 Å) | | 主引用文献 | X-ray Structures of two xanthine inhibitors bound to PEPCK and N-3 modifications of substituted 1,8-Dibenzylxanthines
Bioorg.Med.Chem.Lett., 13, 2003
|
|
1IU2
 
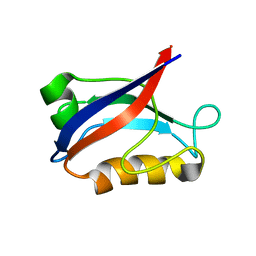 | | The first PDZ domain of PSD-95 | | 分子名称: | PSD-95 | | 著者 | Long, J.-F, Tochio, H, Wang, P, Sala, C, Niethammer, M, Sheng, M, Zhang, M. | | 登録日 | 2002-02-19 | | 公開日 | 2003-03-11 | | 最終更新日 | 2023-12-27 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Supramodular structure and synergistic target binding of the N-terminal tandem PDZ domains of PSD-95
J.MOL.BIOL., 327, 2003
|
|
8H10
 
 | | Structure of SARS-CoV-1 Spike Protein with Engineered x1 Disulfide (S370C and D967C), Locked-2 Conformation | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BILIVERDINE IX ALPHA, ... | | 著者 | Zhang, X, Li, Z, Liu, Y, Wang, J, Fu, L, Wang, P, He, J, Xiong, X. | | 登録日 | 2022-09-30 | | 公開日 | 2022-10-19 | | 最終更新日 | 2024-11-20 | | 実験手法 | ELECTRON MICROSCOPY (2.99 Å) | | 主引用文献 | Disulfide stabilization reveals conserved dynamic features between SARS-CoV-1 and SARS-CoV-2 spikes.
Life Sci Alliance, 6, 2023
|
|
8H13
 
 | | Structure of SARS-CoV-1 Spike Protein with Engineered x2 Disulfide (G400C and V969C), Closed Conformation | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | 著者 | Zhang, X, Li, Z, Liu, Y, Wang, J, Fu, L, Wang, P, He, J, Xiong, X. | | 登録日 | 2022-09-30 | | 公開日 | 2022-10-19 | | 最終更新日 | 2024-11-13 | | 実験手法 | ELECTRON MICROSCOPY (4.05 Å) | | 主引用文献 | Disulfide stabilization reveals conserved dynamic features between SARS-CoV-1 and SARS-CoV-2 spikes.
Life Sci Alliance, 6, 2023
|
|
8H14
 
 | | Structure of SARS-CoV-1 Spike Protein with Engineered x3 Disulfide (D414C and V969C), Locked-1 Conformation | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, LINOLEIC ACID, Spike glycoprotein | | 著者 | Zhang, X, Li, Z, Liu, Y, Wang, J, Fu, L, Wang, P, He, J, Xiong, X. | | 登録日 | 2022-09-30 | | 公開日 | 2022-10-19 | | 最終更新日 | 2024-10-09 | | 実験手法 | ELECTRON MICROSCOPY (3.39 Å) | | 主引用文献 | Disulfide stabilization reveals conserved dynamic features between SARS-CoV-1 and SARS-CoV-2 spikes.
Life Sci Alliance, 6, 2023
|
|
8H16
 
 | | Structure of SARS-CoV-1 Spike Protein (S/native) at pH 5.5, Open Conformation | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | 著者 | Zhang, X, Li, Z, Liu, Y, Wang, J, Fu, L, Wang, P, He, J, Xiong, X. | | 登録日 | 2022-09-30 | | 公開日 | 2022-11-09 | | 最終更新日 | 2024-10-23 | | 実験手法 | ELECTRON MICROSCOPY (3.35534 Å) | | 主引用文献 | Disulfide stabilization reveals conserved dynamic features between SARS-CoV-1 and SARS-CoV-2 spikes.
Life Sci Alliance, 6, 2023
|
|
8H11
 
 | | Structure of SARS-CoV-1 Spike Protein with Engineered x1 Disulfide (S370C and D967C), Closed Conformation | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | 著者 | Zhang, X, Li, Z, Liu, Y, Wang, J, Fu, L, Wang, P, He, J, Xiong, X. | | 登録日 | 2022-09-30 | | 公開日 | 2022-11-09 | | 最終更新日 | 2024-11-06 | | 実験手法 | ELECTRON MICROSCOPY (2.72 Å) | | 主引用文献 | Disulfide stabilization reveals conserved dynamic features between SARS-CoV-1 and SARS-CoV-2 spikes.
Life Sci Alliance, 6, 2023
|
|
8H12
 
 | | Structure of SARS-CoV-1 Spike Protein with Engineered x2 Disulfide (G400C and V969C), Locked-2 Conformation | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | 著者 | Zhang, X, Li, Z, Liu, Y, Wang, J, Fu, L, Wang, P, He, J, Xiong, X. | | 登録日 | 2022-09-30 | | 公開日 | 2022-11-09 | | 最終更新日 | 2024-11-13 | | 実験手法 | ELECTRON MICROSCOPY (3.44681 Å) | | 主引用文献 | Disulfide stabilization reveals conserved dynamic features between SARS-CoV-1 and SARS-CoV-2 spikes.
Life Sci Alliance, 6, 2023
|
|
8H15
 
 | | Structure of SARS-CoV-1 Spike Protein (S/native) at pH 5.5, Closed Conformation | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | 著者 | Zhang, X, Li, Z, Liu, Y, Wang, J, Fu, L, Wang, P, He, J, Xiong, X. | | 登録日 | 2022-09-30 | | 公開日 | 2022-11-09 | | 最終更新日 | 2024-11-20 | | 実験手法 | ELECTRON MICROSCOPY (3.14182 Å) | | 主引用文献 | Disulfide stabilization reveals conserved dynamic features between SARS-CoV-1 and SARS-CoV-2 spikes.
Life Sci Alliance, 6, 2023
|
|
8H0X
 
 | | Structure of SARS-CoV-1 Spike Protein with Engineered x1 Disulfide (S370C and D967C), Locked-1 Conformation | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, BILIVERDINE IX ALPHA, LINOLEIC ACID, ... | | 著者 | Zhang, X, Li, Z, Liu, Y, Wang, J, Fu, L, Wang, P, He, J, Xiong, X. | | 登録日 | 2022-09-30 | | 公開日 | 2022-11-09 | | 最終更新日 | 2024-11-13 | | 実験手法 | ELECTRON MICROSCOPY (2.57 Å) | | 主引用文献 | Disulfide stabilization reveals conserved dynamic features between SARS-CoV-1 and SARS-CoV-2 spikes.
Life Sci Alliance, 6, 2023
|
|
8H0Y
 
 | | Structure of SARS-CoV-1 Spike Protein with Engineered x1 Disulfide (S370C and D967C), Locked-112 Conformation | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, BILIVERDINE IX ALPHA, LINOLEIC ACID, ... | | 著者 | Zhang, X, Li, Z, Liu, Y, Wang, J, Fu, L, Wang, P, He, J, Xiong, X. | | 登録日 | 2022-09-30 | | 公開日 | 2022-11-09 | | 最終更新日 | 2024-11-06 | | 実験手法 | ELECTRON MICROSCOPY (2.85 Å) | | 主引用文献 | Disulfide stabilization reveals conserved dynamic features between SARS-CoV-1 and SARS-CoV-2 spikes.
Life Sci Alliance, 6, 2023
|
|
