7WI6
 
 | | Cryo-EM structure of LY341495/NAM-bound mGlu3 | | 分子名称: | 2-[(1S,2S)-2-carboxycyclopropyl]-3-(9H-xanthen-9-yl)-D-alanine, 2-acetamido-2-deoxy-beta-D-glucopyranose, Metabotropic glutamate receptor 3 | | 著者 | Fang, W, Yang, F, Xu, C.J, Ling, S.L, Lin, L, Zhou, Y.X, Sun, W.J, Wang, X.M, Liu, P, Rondard, P, Pan, S, Pin, J.P, Tian, C.L, Liu, J.F. | | 登録日 | 2022-01-03 | | 公開日 | 2022-03-16 | | 最終更新日 | 2022-07-20 | | 実験手法 | ELECTRON MICROSCOPY (3.71 Å) | | 主引用文献 | Structural basis of the activation of metabotropic glutamate receptor 3.
Cell Res., 32, 2022
|
|
7WIH
 
 | | Cryo-EM structure of LY2794193-bound mGlu3 | | 分子名称: | (1S,2S,4S,5R,6S)-2-amino-4-[(3-methoxybenzene-1-carbonyl)amino]bicyclo[3.1.0]hexane-2,6-dicarboxylic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, Metabotropic glutamate receptor 3 | | 著者 | Fang, W, Yang, F, Xu, C.J, Ling, S.L, Lin, L, Zhou, Y.X, Sun, W.J, Wang, X.M, Liu, P, Rondard, P, Pan, S, Pin, J.P, Tian, C.L, Liu, J.F. | | 登録日 | 2022-01-03 | | 公開日 | 2022-03-16 | | 最終更新日 | 2022-07-20 | | 実験手法 | ELECTRON MICROSCOPY (3.68 Å) | | 主引用文献 | Structural basis of the activation of metabotropic glutamate receptor 3.
Cell Res., 32, 2022
|
|
7WI8
 
 | | Cryo-EM structure of inactive mGlu3 bound to LY341495 | | 分子名称: | 2-[(1S,2S)-2-carboxycyclopropyl]-3-(9H-xanthen-9-yl)-D-alanine, 2-acetamido-2-deoxy-beta-D-glucopyranose, Metabotropic glutamate receptor 3 | | 著者 | Fang, W, Yang, F, Xu, C.J, Ling, S.L, Lin, L, Zhou, Y.X, Sun, W.J, Wang, X.M, Liu, P, Rondard, P, Pan, S, Pin, J.P, Tian, C.L, Liu, J.F. | | 登録日 | 2022-01-03 | | 公開日 | 2022-03-16 | | 最終更新日 | 2022-07-20 | | 実験手法 | ELECTRON MICROSCOPY (4.17 Å) | | 主引用文献 | Structural basis of the activation of metabotropic glutamate receptor 3.
Cell Res., 32, 2022
|
|
8WEX
 
 | |
8EQF
 
 | |
5TRF
 
 | | MDM2 in complex with SAR405838 | | 分子名称: | (2'S,3R,4'S,5'R)-6-chloro-4'-(3-chloro-2-fluorophenyl)-2'-(2,2-dimethylpropyl)-N-(trans-4-hydroxycyclohexyl)-2-oxo-1,2-dihydrospiro[indole-3,3'-pyrrolidine]-5'-carboxamide, E3 ubiquitin-protein ligase Mdm2, GLYCEROL, ... | | 著者 | Meagher, J.L, Stuckey, J.A. | | 登録日 | 2016-10-26 | | 公開日 | 2016-11-09 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | SAR405838: an optimized inhibitor of MDM2-p53 interaction that induces complete and durable tumor regression.
Cancer Res., 74, 2014
|
|
8HTR
 
 | | Crystal structure of Bcl2 in complex with S-9c | | 分子名称: | 4-[4-[(2~{S})-2-(2-chlorophenyl)pyrrolidin-1-yl]phenyl]-~{N}-[3-nitro-4-(oxan-4-ylmethylamino)phenyl]sulfonyl-2-(1~{H}-pyrrolo[2,3-b]pyridin-5-yloxy)benzamide, Apoptosis regulator Bcl-2 | | 著者 | Liu, J, Xu, M, Feng, Y, Liu, Y. | | 登録日 | 2022-12-21 | | 公開日 | 2024-05-15 | | 最終更新日 | 2024-06-05 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Discovery of the Clinical Candidate Sonrotoclax (BGB-11417), a Highly Potent and Selective Inhibitor for Both WT and G101V Mutant Bcl-2.
J.Med.Chem., 67, 2024
|
|
8HTS
 
 | | Crystal structure of Bcl2 in complex with S-10r | | 分子名称: | 4-[2-[(2~{S})-2-(2-cyclopropylphenyl)pyrrolidin-1-yl]-7-azaspiro[3.5]nonan-7-yl]-~{N}-[3-nitro-4-(oxan-4-ylmethylamino)phenyl]sulfonyl-2-(1~{H}-pyrrolo[2,3-b]pyridin-5-yloxy)benzamide, Apoptosis regulator Bcl-2 | | 著者 | Liu, J, Xu, M, Feng, Y, Liu, Y. | | 登録日 | 2022-12-21 | | 公開日 | 2024-05-15 | | 最終更新日 | 2024-06-05 | | 実験手法 | X-RAY DIFFRACTION (1.25 Å) | | 主引用文献 | Discovery of the Clinical Candidate Sonrotoclax (BGB-11417), a Highly Potent and Selective Inhibitor for Both WT and G101V Mutant Bcl-2.
J.Med.Chem., 67, 2024
|
|
6N05
 
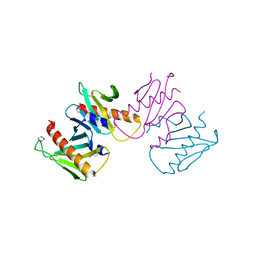 | | Structure of anti-crispr protein, AcrIIC2 | | 分子名称: | AcrIIC2 | | 著者 | Shah, M, Thavalingham, A, Maxwell, K.L, Moraes, T.F. | | 登録日 | 2018-11-06 | | 公開日 | 2019-06-05 | | 最終更新日 | 2020-01-08 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Inhibition of CRISPR-Cas9 ribonucleoprotein complex assembly by anti-CRISPR AcrIIC2.
Nat Commun, 10, 2019
|
|
7U8L
 
 | |
7U8M
 
 | |
7U8J
 
 | |
1A11
 
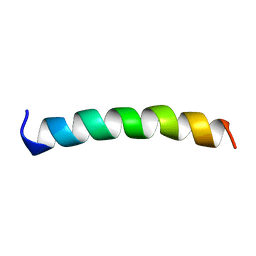 | | NMR STRUCTURE OF MEMBRANE SPANNING SEGMENT 2 OF THE ACETYLCHOLINE RECEPTOR IN DPC MICELLES, 10 STRUCTURES | | 分子名称: | ACETYLCHOLINE RECEPTOR M2 | | 著者 | Gesell, J.J, Sun, W, Montal, M, Opella, S.J. | | 登録日 | 1997-12-19 | | 公開日 | 1998-04-08 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structures of the M2 channel-lining segments from nicotinic acetylcholine and NMDA receptors by NMR spectroscopy.
Nat.Struct.Biol., 6, 1999
|
|
3UDC
 
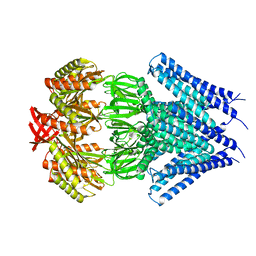 | | Crystal structure of a membrane protein | | 分子名称: | Small-conductance mechanosensitive channel, C-terminal peptide from Small-conductance mechanosensitive channel | | 著者 | Li, W, Ge, J, Yang, M. | | 登録日 | 2011-10-28 | | 公開日 | 2012-10-31 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (3.355 Å) | | 主引用文献 | Structure and molecular mechanism of an anion-selective mechanosensitive channel of small conductance
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3T9N
 
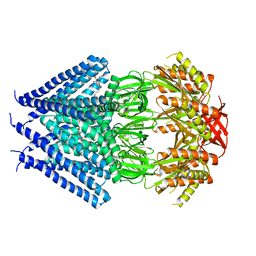 | | Crystal structure of a membrane protein | | 分子名称: | DODECYL-BETA-D-MALTOSIDE, Small-conductance mechanosensitive channel | | 著者 | Yang, M, Zhang, X, Ge, J, Wang, J. | | 登録日 | 2011-08-03 | | 公開日 | 2012-10-31 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (3.456 Å) | | 主引用文献 | Structure and molecular mechanism of an anion-selective mechanosensitive channel of small conductance
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3NCA
 
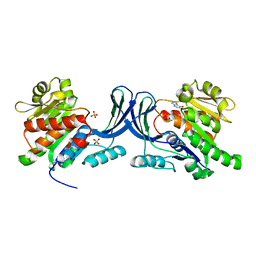 | | X-ray structure of ketohexokinase in complex with a thieno pyridinol compound | | 分子名称: | Ketohexokinase, SULFATE ION, thieno[3,2-b]pyridin-7-ol | | 著者 | Abad, M.C, Gibbs, A.C, Spurlino, J.C. | | 登録日 | 2010-06-04 | | 公開日 | 2010-12-22 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.602 Å) | | 主引用文献 | Electron density guided fragment-based lead discovery of ketohexokinase inhibitors.
J.Med.Chem., 53, 2010
|
|
7EOD
 
 | | MITF HLHLZ Delta AKE | | 分子名称: | GLYCEROL, Isoform M1 of Microphthalmia-associated transcription factor | | 著者 | Li, P, Liu, Z, Fang, P, Wang, J. | | 登録日 | 2021-04-22 | | 公開日 | 2022-04-27 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | A unique hyperdynamic dimer interface permits small molecule perturbation of the melanoma oncoprotein MITF for melanoma therapy.
Cell Res., 33, 2023
|
|
3O0I
 
 | |
3O2F
 
 | |
3RO4
 
 | |
3KMP
 
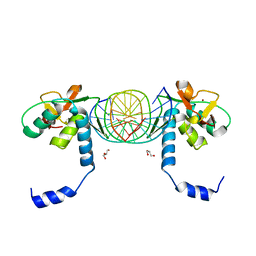 | | Crystal Structure of SMAD1-MH1/DNA complex | | 分子名称: | 5'-D(P*AP*TP*CP*AP*GP*TP*CP*TP*AP*GP*AP*CP*AP*TP*A)-3', 5'-D(P*GP*TP*AP*TP*GP*TP*CP*TP*AP*GP*AP*CP*TP*GP*A)-3', GLYCEROL, ... | | 著者 | Baburajendran, N, Palasingam, P, Narasimhan, K, Jauch, R, Kolatkar, P.R. | | 登録日 | 2009-11-11 | | 公開日 | 2010-02-23 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Structure of Smad1 MH1/DNA complex reveals distinctive rearrangements of BMP and TGF-beta effectors.
Nucleic Acids Res., 38, 2010
|
|
3NBW
 
 | | X-ray structure of ketohexokinase in complex with a pyrazole compound | | 分子名称: | 5-amino-3-(methylsulfanyl)-1-phenyl-1H-pyrazole-4-carbonitrile, GLYCEROL, Ketohexokinase, ... | | 著者 | Abad, M.C, Gibbs, A.C, Spurlino, J.C. | | 登録日 | 2010-06-04 | | 公開日 | 2010-12-22 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.341 Å) | | 主引用文献 | Electron density guided fragment-based lead discovery of ketohexokinase inhibitors.
J.Med.Chem., 53, 2010
|
|
3NC2
 
 | |
3NC9
 
 | |
3NBV
 
 | |
