2EHD
 
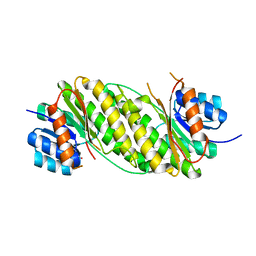 | | Crystal Structure Analysis of Oxidoreductase | | 分子名称: | COBALT (II) ION, Oxidoreductase, short-chain dehydrogenase/reductase family | | 著者 | Kamiya, N, Hikima, T, Ebihara, A, Inoue, Y, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | 登録日 | 2007-03-06 | | 公開日 | 2008-03-11 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Crystal structure analysis of putative oxidoreductase from Thermus thermophilus HB8
to be published
|
|
2YR5
 
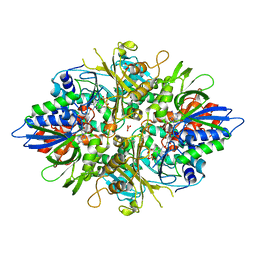 | | Crystal structure of L-phenylalanine oxidase from Psuedomonas sp.P501 | | 分子名称: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, Pro-enzyme of L-phenylalanine oxidase, ... | | 著者 | Ida, K, Kurabayashi, M, Suguro, M, Hikima, T, Yamamoto, M, Suzuki, H. | | 登録日 | 2007-04-02 | | 公開日 | 2008-04-15 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.25 Å) | | 主引用文献 | Structural basis of proteolytic activation of L-phenylalanine oxidase from Pseudomonas sp. P-501.
J.Biol.Chem., 283, 2008
|
|
5T79
 
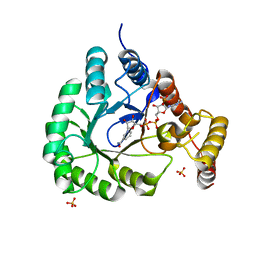 | | X-Ray Crystal Structure of a Novel Aldo-keto Reductases for the Biocatalytic Conversion of 3-hydroxybutanal to 1,3-butanediol | | 分子名称: | Aldo-keto Reductase, OXIDOREDUCTASE, CHLORIDE ION, ... | | 著者 | Brunzelle, J.S, Wawrzak, Z, Evdokimova, E, Kudritska, M, Savchenko, A, Yakunin, A.F, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2016-09-02 | | 公開日 | 2017-02-15 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.86 Å) | | 主引用文献 | Structural and biochemical studies of novel aldo-keto reductases for the biocatalytic conversion of 3-hydroxybutanal to 1,3-butanediol.
Appl. Environ. Microbiol., 2017
|
|
3ERP
 
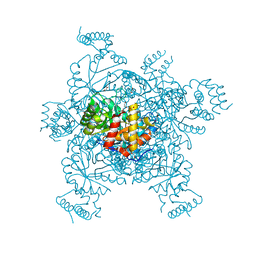 | | Structure of IDP01002, a putative oxidoreductase from and essential gene of Salmonella typhimurium | | 分子名称: | 1,2-ETHANEDIOL, CACODYLATE ION, CHLORIDE ION, ... | | 著者 | Singer, A.U, Minasov, G, Evdokimova, E, Brunzelle, J.S, Kudritska, M, Edwards, A.M, Anderson, W.F, Savchenko, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2008-10-02 | | 公開日 | 2008-11-04 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.55 Å) | | 主引用文献 | Structural and biochemical studies of novel aldo-keto reductases for the biocatalytic conversion of 3-hydroxybutanal to 1,3-butanediol.
Appl.Environ.Microbiol., 2017
|
|
2KQK
 
 | | Solution structure of apo-IscU(D39A) | | 分子名称: | NifU-like protein | | 著者 | Kim, J.H, Fuzery, A.K, Tonelli, M, Vickery, L.E, Markley, J.L. | | 登録日 | 2009-11-10 | | 公開日 | 2010-11-17 | | 最終更新日 | 2024-05-08 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Three-Dimensional Structure and Determinants of Stability of the Iron-Sulfur Cluster Scaffold Protein IscU from Escherichia coli.
Biochemistry, 51, 2012
|
|
7SHG
 
 | |
3JUJ
 
 | |
3JUK
 
 | |
8IVU
 
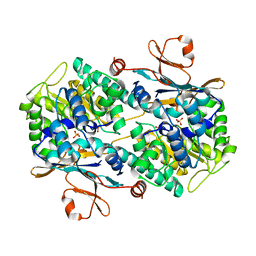 | | Crystal Structure of Human NAMPT in complex with A4276 | | 分子名称: | N-[[4-(6-methyl-1,3-benzoxazol-2-yl)phenyl]methyl]pyridine-3-carboxamide, Nicotinamide phosphoribosyltransferase, PHOSPHATE ION | | 著者 | Kang, B.G, Cha, S.S. | | 登録日 | 2023-03-28 | | 公開日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.09000921 Å) | | 主引用文献 | Discovery of a novel NAMPT inhibitor that selectively targets NAPRT-deficient EMT-subtype cancer cells and alleviates chemotherapy-induced peripheral neuropathy.
Theranostics, 13, 2023
|
|
5GUY
 
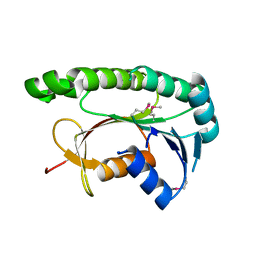 | | Structural and functional study of chuY from E. coli CFT073 strain | | 分子名称: | Uncharacterized protein chuY | | 著者 | Kim, H, Choi, J, HA, S.C, Chaurasia, A.K, Kim, D, Kim, K.K. | | 登録日 | 2016-08-31 | | 公開日 | 2017-03-08 | | 実験手法 | X-RAY DIFFRACTION (2.399 Å) | | 主引用文献 | Structural and functional study of ChuY from Escherichia coli strain CFT073
Biochem. Biophys. Res. Commun., 482, 2017
|
|
8ECZ
 
 | | Bovine Fab 4C1 | | 分子名称: | 4C1 Fab heavy chain, 4C1 Fab light chain, PHOSPHATE ION | | 著者 | Stanfield, R.L, Wilson, I.A. | | 登録日 | 2022-09-02 | | 公開日 | 2023-09-20 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.82 Å) | | 主引用文献 | The smallest functional antibody fragment: Ultralong CDR H3 antibody knob regions potently neutralize SARS-CoV-2.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8ECV
 
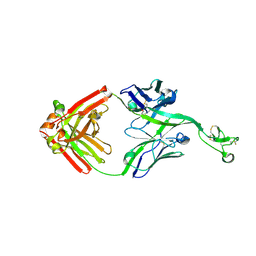 | | Bovine Fab 2F12 | | 分子名称: | 2F12 Fab Heavy chain, 2F12 Fab Light chain | | 著者 | Stanfield, R.L, Wilson, I.A. | | 登録日 | 2022-09-02 | | 公開日 | 2023-09-20 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.81 Å) | | 主引用文献 | The smallest functional antibody fragment: Ultralong CDR H3 antibody knob regions potently neutralize SARS-CoV-2.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8ECQ
 
 | | Bovine Fab 2G3 | | 分子名称: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2G3 Fab Heavy chain, 2G3 Fab Light chain, ... | | 著者 | Stanfield, R.L, Wilson, I.A. | | 登録日 | 2022-09-02 | | 公開日 | 2023-09-20 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | The smallest functional antibody fragment: Ultralong CDR H3 antibody knob regions potently neutralize SARS-CoV-2.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8ED1
 
 | | Bovine Fab 5C1 | | 分子名称: | 5C1 Fab heavy chain, 5C1 Fab light chain, GLYCEROL, ... | | 著者 | Stanfield, R.L, Wilson, I.A. | | 登録日 | 2022-09-02 | | 公開日 | 2023-09-20 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.31 Å) | | 主引用文献 | The smallest functional antibody fragment: Ultralong CDR H3 antibody knob regions potently neutralize SARS-CoV-2.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8EDF
 
 | |
5DNY
 
 | | Structure of the ATPrS-Mre11/Rad50-DNA complex | | 分子名称: | DNA (27-MER), DNA double-strand break repair Rad50 ATPase,DNA double-strand break repair Rad50 ATPase, DNA double-strand break repair protein Mre11, ... | | 著者 | Liu, Y. | | 登録日 | 2015-09-10 | | 公開日 | 2016-02-24 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (3.11 Å) | | 主引用文献 | ATP-dependent DNA binding, unwinding, and resection by the Mre11/Rad50 complex.
Embo J., 35, 2016
|
|
5F3W
 
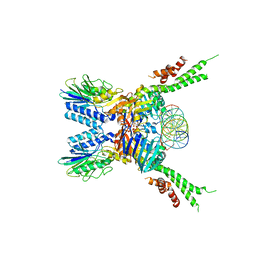 | | Structure of the ATPrS-Mre11/Rad50-DNA complex | | 分子名称: | 27-MER DNA, DNA double-strand break repair Rad50 ATPase,DNA double-strand break repair Rad50 ATPase, DNA double-strand break repair protein Mre11, ... | | 著者 | Liu, Y. | | 登録日 | 2015-12-03 | | 公開日 | 2016-03-02 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (3.11 Å) | | 主引用文献 | ATP-dependent DNA binding, unwinding, and resection by the Mre11/Rad50 complex
Embo J., 35, 2016
|
|
1DLF
 
 | | HIGH RESOLUTION CRYSTAL STRUCTURE OF THE FV FRAGMENT FROM AN ANTI-DANSYL SWITCH VARIANT ANTIBODY IGG2A(S) CRYSTALLIZED AT PH 5.25 | | 分子名称: | ANTI-DANSYL IMMUNOGLOBULIN IGG2A(S), SULFATE ION | | 著者 | Nakasako, M, Takahashi, H, Shimada, I, Arata, Y. | | 登録日 | 1998-07-14 | | 公開日 | 1999-07-26 | | 最終更新日 | 2023-08-09 | | 実験手法 | X-RAY DIFFRACTION (1.45 Å) | | 主引用文献 | The pH-dependent structural variation of complementarity-determining region H3 in the crystal structures of the Fv fragment from an anti-dansyl monoclonal antibody.
J.Mol.Biol., 291, 1999
|
|
2DLF
 
 | | HIGH RESOLUTION CRYSTAL STRUCTURE OF THE FV FRAGMENT FROM AN ANTI-DANSYL SWITCH VARIANT ANTIBODY IGG2A(S) CRYSTALLIZED AT PH 6.75 | | 分子名称: | PROTEIN (ANTI-DANSYL IMMUNOGLOBULIN IGG2A(S) (HEAVY CHAIN)), PROTEIN (ANTI-DANSYL IMMUNOGLOBULIN IGG2A(S)-KAPPA (LIGHT CHAIN)), SULFATE ION | | 著者 | Nakasako, M, Takahashi, H, Shimada, I, Arata, Y. | | 登録日 | 1998-12-17 | | 公開日 | 1999-12-17 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (1.55 Å) | | 主引用文献 | The pH-dependent structural variation of complementarity-determining region H3 in the crystal structures of the Fv fragment from an anti-dansyl monoclonal antibody.
J.Mol.Biol., 291, 1999
|
|
2L4X
 
 | | Solution Structure of apo-IscU(WT) | | 分子名称: | Iron-sulfur cluster assembly scaffold protein | | 著者 | Kim, J.H, Tonelli, M, Markley, J.L. | | 登録日 | 2010-10-19 | | 公開日 | 2011-12-07 | | 最終更新日 | 2024-05-01 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Three-Dimensional Structure and Determinants of Stability of the Iron-Sulfur Cluster Scaffold Protein IscU from Escherichia coli.
Biochemistry, 51, 2012
|
|
6JTT
 
 | | MHETase in complex with BHET | | 分子名称: | 4-(2-hydroxyethyloxycarbonyl)benzoic acid, CALCIUM ION, Mono(2-hydroxyethyl) terephthalate hydrolase, ... | | 著者 | Sagong, H.-Y, Seo, H, Kim, K.-J. | | 登録日 | 2019-04-12 | | 公開日 | 2020-04-15 | | 最終更新日 | 2020-10-28 | | 実験手法 | X-RAY DIFFRACTION (2.51 Å) | | 主引用文献 | Decomposition of PET film by MHETase using Exo-PETase function
Acs Catalysis, 10, 2020
|
|
4UBC
 
 | | DNA polymerase beta substrate complex with a templating cytosine and incoming 8-oxodGTP, 0 s | | 分子名称: | 5'-D(*CP*CP*GP*AP*CP*CP*GP*CP*GP*CP*AP*TP*CP*AP*GP*C)-3', 5'-D(*GP*CP*TP*GP*AP*TP*GP*CP*GP*C)-3', 5'-D(P*GP*TP*CP*GP*G)-3', ... | | 著者 | Freudenthal, B.D, Wilson, S.H, Beard, W.A. | | 登録日 | 2014-08-12 | | 公開日 | 2014-11-12 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Uncovering the polymerase-induced cytotoxicity of an oxidized nucleotide.
Nature, 517, 2015
|
|
4UAZ
 
 | | DNA polymerase beta reactant complex with a templating adenine and incoming 8-oxodGTP, 20 s | | 分子名称: | 5'-D(*CP*CP*GP*AP*CP*AP*GP*CP*GP*CP*AP*TP*CP*AP*GP*C)-3', 5'-D(*GP*CP*TP*GP*AP*TP*GP*CP*GP*CP*(8OG))-3', 5'-D(P*GP*TP*CP*GP*G)-3', ... | | 著者 | Freudenthal, B.D, Wilson, S.H, Beard, W.A. | | 登録日 | 2014-08-11 | | 公開日 | 2014-11-12 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.88 Å) | | 主引用文献 | Uncovering the polymerase-induced cytotoxicity of an oxidized nucleotide.
Nature, 517, 2015
|
|
4UB3
 
 | | DNA polymerase beta closed product complex with a templating cytosine and 8-oxodGMP, 60 s | | 分子名称: | 5'-D(*CP*CP*GP*AP*CP*CP*GP*CP*GP*CP*AP*TP*CP*AP*GP*C)-3', 5'-D(*GP*CP*TP*GP*AP*TP*GP*CP*GP*CP*(8OG))-3', 5'-D(P*GP*TP*CP*GP*G)-3', ... | | 著者 | Freudenthal, B.D, Wilson, S.H, Beard, W.A. | | 登録日 | 2014-08-11 | | 公開日 | 2014-11-12 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.06 Å) | | 主引用文献 | Uncovering the polymerase-induced cytotoxicity of an oxidized nucleotide.
Nature, 517, 2015
|
|
4UBB
 
 | | DNA polymerase beta reactant complex with a templating cytosine and incoming 8-oxodGTP, 40 s | | 分子名称: | 5'-D(*CP*CP*GP*AP*CP*CP*GP*CP*GP*CP*AP*TP*CP*AP*GP*C)-3', 5'-D(*GP*CP*TP*GP*AP*TP*GP*CP*GP*CP*(8OG))-3', 5'-D(P*GP*TP*CP*GP*G)-3', ... | | 著者 | Freudenthal, B.D, Wilson, S.H, Beard, W.A. | | 登録日 | 2014-08-12 | | 公開日 | 2014-11-12 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Uncovering the polymerase-induced cytotoxicity of an oxidized nucleotide.
Nature, 517, 2015
|
|
