4FOM
 
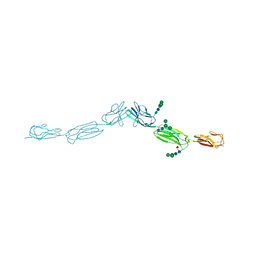 | | Crystal structure of human nectin-3 full ectodomain (D1-D3) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Poliovirus receptor-related protein 3, alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Harrison, O.J, Jin, X, Brasch, J, Shapiro, L. | | 登録日 | 2012-06-20 | | 公開日 | 2012-08-22 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (3.93 Å) | | 主引用文献 | Nectin ectodomain structures reveal a canonical adhesive interface.
Nat.Struct.Mol.Biol., 19, 2012
|
|
4FRW
 
 | |
3UL8
 
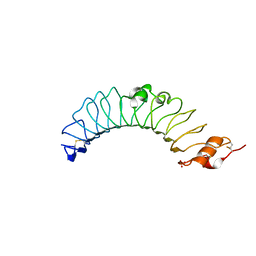 | | Crystal structure of the TV3 mutant V134L | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, SULFATE ION, Toll-like receptor 4, ... | | 著者 | Kim, H.J, Cheong, H.K, Jeon, Y.H. | | 登録日 | 2011-11-10 | | 公開日 | 2012-04-04 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Structure-Based Rational Design of a Toll-like Receptor 4 (TLR4) Decoy Receptor with High Binding Affinity for a Target Protein.
Plos One, 7, 2012
|
|
3ULA
 
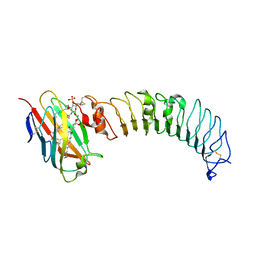 | | Crystal structure of the TV3 mutant F63W-MD-2-Eritoran complex | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 3-O-DECYL-2-DEOXY-6-O-{2-DEOXY-3-O-[(3R)-3-METHOXYDECYL]-6-O-METHYL-2-[(11Z)-OCTADEC-11-ENOYLAMINO]-4-O-PHOSPHONO-BETA-D-GLUCOPYRANOSYL}-2-[(3-OXOTETRADECANOYL)AMINO]-1-O-PHOSPHONO-ALPHA-D-GLUCOPYRANOSE, Lymphocyte antigen 96, ... | | 著者 | Kim, H.J, Cheong, H.K, Jeon, Y.H. | | 登録日 | 2011-11-10 | | 公開日 | 2012-04-04 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (3.6 Å) | | 主引用文献 | Structure-Based Rational Design of a Toll-like Receptor 4 (TLR4) Decoy Receptor with High Binding Affinity for a Target Protein.
Plos One, 7, 2012
|
|
3UL7
 
 | | Crystal structure of the TV3 mutant F63W | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, SULFATE ION, Toll-like receptor 4, ... | | 著者 | Kim, H.J, Cheong, H.K, Jeon, Y.H. | | 登録日 | 2011-11-10 | | 公開日 | 2012-04-04 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.37 Å) | | 主引用文献 | Structure-Based Rational Design of a Toll-like Receptor 4 (TLR4) Decoy Receptor with High Binding Affinity for a Target Protein.
Plos One, 7, 2012
|
|
3UL9
 
 | | structure of the TV3 mutant M41E | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, SULFATE ION, Toll-like receptor 4, ... | | 著者 | Kim, H.J, Cheong, H.K, Jeon, Y.H. | | 登録日 | 2011-11-10 | | 公開日 | 2012-05-23 | | 最終更新日 | 2024-10-09 | | 実験手法 | X-RAY DIFFRACTION (2.45 Å) | | 主引用文献 | Structure-Based Rational Design of a Toll-like Receptor 4 (TLR4) Decoy Receptor with High Binding Affinity for a Target Protein.
Plos One, 7, 2012
|
|
3Q2N
 
 | |
6M3N
 
 | | Solution structure of anti-CRISPR AcrIF7 | | 分子名称: | anti-CRIPSR AcrIF7 | | 著者 | Kim, I, An, S.Y, Koo, J, Bae, E, Suh, J.Y. | | 登録日 | 2020-03-04 | | 公開日 | 2020-08-26 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structural and mechanistic insights into the CRISPR inhibition of AcrIF7.
Nucleic Acids Res., 48, 2020
|
|
3Q2L
 
 | |
3Q2V
 
 | | Crystal structure of mouse E-cadherin ectodomain | | 分子名称: | CALCIUM ION, Cadherin-1, MANGANESE (II) ION, ... | | 著者 | Jin, X, Harrison, O.J, Shapiro, L. | | 登録日 | 2010-12-20 | | 公開日 | 2011-04-06 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (3.4 Å) | | 主引用文献 | The extracellular architecture of adherens junctions revealed by crystal structures of type I cadherins.
Structure, 19, 2011
|
|
3Q2W
 
 | | Crystal structure of mouse N-cadherin ectodomain | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | 著者 | Jin, X, Shapiro, L. | | 登録日 | 2010-12-20 | | 公開日 | 2011-02-23 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (3.2 Å) | | 主引用文献 | The extracellular architecture of adherens junctions revealed by crystal structures of type I cadherins.
Structure, 19, 2011
|
|
3RFS
 
 | | Design of a binding scaffold based on variable lymphocyte receptors of jawless vertebrates by module engineering | | 分子名称: | Internalin B, repeat modules, Variable lymphocyte receptor B, ... | | 著者 | Kim, H.J, Cheong, H.K, Jeon, Y.H. | | 登録日 | 2011-04-06 | | 公開日 | 2012-03-14 | | 最終更新日 | 2017-08-16 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Design of a binding scaffold based on variable lymphocyte receptors of jawless vertebrates by module engineering
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3RFJ
 
 | | Design of a binding scaffold based on variable lymphocyte receptors of jawless vertebrates by module engineering | | 分子名称: | Internalin B, repeat modules, Variable lymphocyte receptor, ... | | 著者 | Kim, H.J, Cheong, H.K, Jeon, Y.H. | | 登録日 | 2011-04-06 | | 公開日 | 2012-03-14 | | 最終更新日 | 2017-08-16 | | 実験手法 | X-RAY DIFFRACTION (1.78 Å) | | 主引用文献 | Design of a binding scaffold based on variable lymphocyte receptors of jawless vertebrates by module engineering
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
8W5J
 
 | | Cryo-EM structure of the yeast TOM core complex (from TOM-TIM23 complex) | | 分子名称: | (2R)-3-{[(S)-(2-aminoethoxy)(hydroxy)phosphoryl]oxy}-2-(tetradecanoyloxy)propyl tetradecanoate, Mitochondrial import receptor subunit TOM22, Mitochondrial import receptor subunit TOM40, ... | | 著者 | Wang, Q, Guan, Z.Y, Zhuang, J.J, Huang, R, Yin, P. | | 登録日 | 2023-08-27 | | 公開日 | 2024-02-28 | | 実験手法 | ELECTRON MICROSCOPY (4.4 Å) | | 主引用文献 | The architecture of substrate-engaged TOM-TIM23 supercomplex reveals preprotein proximity sites for mitochondrial protein translocation.
Cell Discov, 10, 2024
|
|
8W5K
 
 | | Cryo-EM structure of the yeast TOM core complex crosslinked by BS3 (from TOM-TIM23 complex) | | 分子名称: | (2R)-3-{[(S)-(2-aminoethoxy)(hydroxy)phosphoryl]oxy}-2-(tetradecanoyloxy)propyl tetradecanoate, Mitochondrial import receptor subunit TOM22, Mitochondrial import receptor subunit TOM40, ... | | 著者 | Wang, Q, Guan, Z.Y, Zhuang, J.J, Huang, R, Yin, P. | | 登録日 | 2023-08-27 | | 公開日 | 2024-03-13 | | 実験手法 | ELECTRON MICROSCOPY (3.6 Å) | | 主引用文献 | The architecture of substrate-engaged TOM-TIM23 supercomplex reveals preprotein proximity sites for mitochondrial protein translocation.
Cell Discov, 10, 2024
|
|
6KGY
 
 | |
6KMH
 
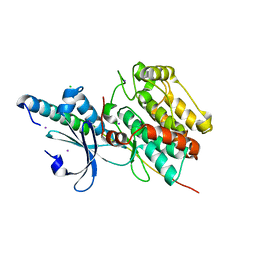 | | The crystal structure of CASK/Mint1 complex | | 分子名称: | Amyloid-beta A4 precursor protein-binding family A member 1, CHLORIDE ION, IODIDE ION, ... | | 著者 | Li, W, Feng, W. | | 登録日 | 2019-07-31 | | 公開日 | 2020-08-05 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | CASK modulates the assembly and function of the Mint1/Munc18-1 complex to regulate insulin secretion.
Cell Discov, 6, 2020
|
|
5ZNX
 
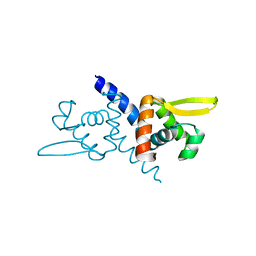 | | Crystal structure of CM14-treated HlyU from Vibrio vulnificus | | 分子名称: | Transcriptional activator | | 著者 | Park, N, Kim, S, Jo, I, Ahn, J, Hong, S, Jeong, S, Baek, Y. | | 登録日 | 2018-04-11 | | 公開日 | 2019-04-10 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.114 Å) | | 主引用文献 | Small-molecule inhibitor of HlyU attenuates virulence of Vibrio species.
Sci Rep, 9, 2019
|
|
5X41
 
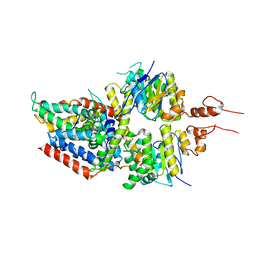 | | 3.5A resolution structure of a cobalt energy-coupling factor transporter using LCP method-CbiMQO | | 分子名称: | Cobalt ABC transporter ATP-binding protein, Cobalt transport protein CbiM, Uncharacterized protein CbiQ | | 著者 | Bao, Z, Qi, X, Zhao, W, Li, D, Zhang, P. | | 登録日 | 2017-02-09 | | 公開日 | 2017-04-19 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (3.47 Å) | | 主引用文献 | Structure and mechanism of a group-I cobalt energy coupling factor transporter
Cell Res., 27, 2017
|
|
5X40
 
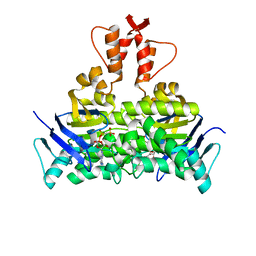 | | Structure of a CbiO dimer bound with AMPPCP | | 分子名称: | Cobalt ABC transporter ATP-binding protein, MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER | | 著者 | Bao, Z, Qi, X, Wang, J, Zhang, P. | | 登録日 | 2017-02-09 | | 公開日 | 2017-04-05 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (1.45 Å) | | 主引用文献 | Structure and mechanism of a group-I cobalt energy coupling factor transporter
Cell Res., 27, 2017
|
|
5X3X
 
 | | 2.8A resolution structure of a cobalt energy-coupling factor transporter-CbiMQO | | 分子名称: | Cobalt ABC transporter ATP-binding protein, Cobalt transport protein CbiM, Uncharacterized protein CbiQ | | 著者 | Bao, Z, Qi, X, Wang, J, Zhang, P. | | 登録日 | 2017-02-09 | | 公開日 | 2017-04-05 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (2.788 Å) | | 主引用文献 | Structure and mechanism of a group-I cobalt energy coupling factor transporter
Cell Res., 27, 2017
|
|
3H1J
 
 | | Stigmatellin-bound cytochrome bc1 complex from chicken | | 分子名称: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, CARDIOLIPIN, Coenzyme Q10, ... | | 著者 | Zhang, Z, Huang, L, Shulmeister, V.M, Chi, Y.-I, Kim, K.K, Hung, L.-W, Crofts, A.R, Berry, E.A, Kim, S.-H. | | 登録日 | 2009-04-12 | | 公開日 | 2009-04-28 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Electron Transfer by Domain Movement in Cytochrome Bc1
Nature, 392, 1998
|
|
2BCC
 
 | | STIGMATELLIN-BOUND CYTOCHROME BC1 COMPLEX FROM CHICKEN | | 分子名称: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, FE2/S2 (INORGANIC) CLUSTER, PROTOPORPHYRIN IX CONTAINING FE, ... | | 著者 | Zhang, Z, Huang, L, Shulmeister, V.M, Chi, Y.I, Kim, K.K, Hung, L.W, Crofts, A.R, Berry, E.A, Kim, S.H. | | 登録日 | 1998-09-18 | | 公開日 | 1999-08-02 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (3.5 Å) | | 主引用文献 | Electron Transfer by Domain Movement in Cytochrome Bc1
Nature, 392, 1998
|
|
7TVW
 
 | | Crystal structure of Arabidopsis thaliana DLK2 | | 分子名称: | Alpha/beta-Hydrolases superfamily protein | | 著者 | Burger, M, Chory, J. | | 登録日 | 2022-02-06 | | 公開日 | 2022-09-28 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.48 Å) | | 主引用文献 | Crystal structure of Arabidopsis DWARF14-LIKE2 (DLK2) reveals a distinct substrate binding pocket architecture.
Plant Direct, 6, 2022
|
|
7DOG
 
 | |
