7RGE
 
 | | Crystal structure of phosphoadenylyl-sulfate (PAPS) reductase from Candida auris, phosphate complex | | 分子名称: | 3'-phosphoadenylylsulfate reductase, GLYCEROL, PHOSPHATE ION | | 著者 | Stogios, P.J, Evdokimova, E, Di Leo, R, Savchenko, A, Joachimiak, A, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2021-07-15 | | 公開日 | 2021-07-28 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.38 Å) | | 主引用文献 | Crystal structure of phosphoadenylyl-sulfate (PAPS) reductase from Candida auris, phosphate complex
To Be Published
|
|
7RH8
 
 | | Crystal structure of Fur1p from Candida albicans, in complex with UTP | | 分子名称: | URIDINE 5'-TRIPHOSPHATE, Uracil phosphoribosyltransferase | | 著者 | Stogios, P.J, Skarina, T, Kim, Y, Di Leo, R, Savchenko, A, Joachimiak, A, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2021-07-16 | | 公開日 | 2021-07-28 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.69 Å) | | 主引用文献 | Crystal structure of Fur1p from Candida albicans, in complex with UTP
To Be Published
|
|
7RJ1
 
 | | Crystal structure of Aro7p chorismate mutase from Candida albicans, complex with L-Trp | | 分子名称: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Chorismate mutase, ... | | 著者 | Stogios, P.J, Evdokimova, E, Tan, K, Di Leo, R, Savchenko, A, Joachimiak, A, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2021-07-20 | | 公開日 | 2021-07-28 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.16 Å) | | 主引用文献 | Crystal structure of Aro7p chorismate mutase from Candida albicans, complex with L-Trp
To Be Published
|
|
7REU
 
 | | Crystal structure of Aro4p, 3-deoxy-D-arabino-heptulosonate-7-phosphate (DAHP) synthase from Candida auris, L-Tyr complex | | 分子名称: | 3-deoxy-D-arabino-heptulosonate-7-phosphate (DAHP) synthase, CHLORIDE ION, GLYCEROL, ... | | 著者 | Stogios, P.J, Evdokimova, E, Tan, K, Di Leo, R, Savchenko, A, Joachimiak, A, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2021-07-13 | | 公開日 | 2021-07-28 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.79 Å) | | 主引用文献 | Crystal structure of Aro4p, 3-deoxy-D-arabino-heptulosonate-7-phosphate (DAHP) synthase from Candida auris, L-Tyr complex
To Be Published
|
|
7RLL
 
 | | Crystal structure of ARF3 from Candida albicans in complex with guanosine-3'-monophosphate-5'-diphosphate | | 分子名称: | Arf3p, GUANOSINE-3'-MONOPHOSPHATE-5'-DIPHOSPHATE, MAGNESIUM ION | | 著者 | Stogios, P.J, Michalska, K, Evdokimova, E, Di Leo, R, Savchenko, A, Joachimiak, A, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2021-07-25 | | 公開日 | 2021-08-11 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Crystal structure of ARF3 from Candida albicans in complex with guanosine-3'-monophosphate-5'-diphosphate
To Be Published
|
|
7RQG
 
 | | Crystal structure of the Nsp3 Y3 domain from SARS-CoV-2 | | 分子名称: | Non-structural protein 3 | | 著者 | Stogios, P.J, Skarina, T, Chang, C, Di Leo, R, Savchenko, A, Joachimiak, A, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2021-08-06 | | 公開日 | 2021-08-18 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.17 Å) | | 主引用文献 | Crystal structure of the Nsp3 Y3 domain from SARS-CoV-2
To Be Published
|
|
6VOP
 
 | | Crystal structure of YgbL, a putative aldolase/epimerase/decarboxylase from Escherichia coli | | 分子名称: | Aldolase | | 著者 | Stogios, P.J, Evdokimova, E, Di Leo, R, Savchenko, A, Joachimiak, A, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2020-01-31 | | 公開日 | 2020-02-19 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.65 Å) | | 主引用文献 | Crystal structure of YgbL, a putative aldolase/epimerase/decarboxylase from Escherichia coli
To Be Published
|
|
6VOQ
 
 | | Crystal structure of YgbL, a putative aldolase/epimerase/decarboxylase from Klebsiella pneumoniae | | 分子名称: | Aldolase, CHLORIDE ION, ZINC ION | | 著者 | Stogios, P.J, Evdokimova, E, McChesney, C, Di Leo, R, Savchenko, A, Joachimiak, A, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2020-01-31 | | 公開日 | 2020-02-19 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.49 Å) | | 主引用文献 | Crystal structure of YgbL, a putative aldolase/epimerase/decarboxylase from Klebsiella pneumoniae
To Be Published
|
|
6VTV
 
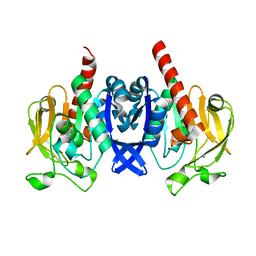 | | Crystal structure of PuuD gamma-glutamyl-gamma-aminobutyrate hydrolase from E. coli | | 分子名称: | Gamma-glutamyl-gamma-aminobutyrate hydrolase PuuD, MANGANESE (II) ION | | 著者 | Stogios, P.J, EVDOKIMOVA, E, DI LEO, R, SAVCHENKO, A, JOACHIMIAK, A, SATCHELL, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2020-02-13 | | 公開日 | 2020-02-26 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.06 Å) | | 主引用文献 | PuuD gamma-glutamyl-gamma-aminobutyrate hydrolase
To Be Published
|
|
4WZ0
 
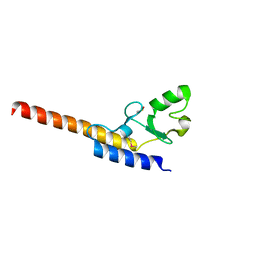 | | Crystal structure of U-box 1 of LubX / LegU2 / Lpp2887 from Legionella pneumophila str. Paris | | 分子名称: | E3 ubiquitin-protein ligase LubX | | 著者 | Stogios, P.J, Quaile, A.T, Skarina, T, Stein, A, Di Leo, R, Yim, V, Savchenko, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2014-11-18 | | 公開日 | 2015-01-14 | | 最終更新日 | 2019-12-04 | | 実験手法 | X-RAY DIFFRACTION (1.954 Å) | | 主引用文献 | Molecular Characterization of LubX: Functional Divergence of the U-Box Fold by Legionella pneumophila.
Structure, 23, 2015
|
|
6VU7
 
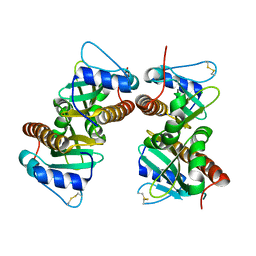 | | Crystal structure of YbjN, a putative transcription regulator from E. coli | | 分子名称: | CHLORIDE ION, YbjN protein | | 著者 | Stogios, P.J, Evdokimova, E, Di Leo, R, Savchenko, A, Joachimiak, A, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2020-02-14 | | 公開日 | 2020-03-11 | | 最終更新日 | 2024-10-09 | | 実験手法 | X-RAY DIFFRACTION (2.59 Å) | | 主引用文献 | Crystal structure of YbjN, a putative transcription regulator from E. coli
To Be Published
|
|
4I3F
 
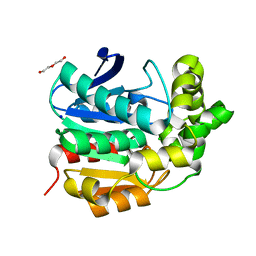 | | Crystal structure of serine hydrolase CCSP0084 from the polyaromatic hydrocarbon (PAH)-degrading bacterium Cycloclasticus zankles | | 分子名称: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, SODIUM ION, ... | | 著者 | Stogios, P.J, Xu, X, Dong, A, Cui, H, Alcaide, M, Tornes, J, Gertler, C, Yakimov, M.M, Golyshin, P.N, Ferrer, M, Savchenko, A. | | 登録日 | 2012-11-26 | | 公開日 | 2013-06-26 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.69 Å) | | 主引用文献 | Single residues dictate the co-evolution of dual esterases: MCP hydrolases from the alpha / beta hydrolase family.
Biochem.J., 454, 2013
|
|
7TOK
 
 | | Crystal structure of the CBM domain of carbohydrate esterase FjoAcXE | | 分子名称: | Acetylxylan esterase I | | 著者 | Stogios, P.J, Skarina, T, Di Leo, R, Jurak, E, Master, E. | | 登録日 | 2022-01-24 | | 公開日 | 2022-04-13 | | 最終更新日 | 2022-11-02 | | 実験手法 | X-RAY DIFFRACTION (2.45 Å) | | 主引用文献 | Elucidating Sequence and Structural Determinants of Carbohydrate Esterases for Complete Deacetylation of Substituted Xylans.
Molecules, 27, 2022
|
|
7TOG
 
 | | Crystal structure of carbohydrate esterase PbeAcXE, apoenzyme | | 分子名称: | SGNH hydrolase | | 著者 | Stogios, P.J, Skarina, T, Di Leo, R, Jurak, E, Master, E. | | 登録日 | 2022-01-24 | | 公開日 | 2022-04-13 | | 最終更新日 | 2022-11-02 | | 実験手法 | X-RAY DIFFRACTION (1.35 Å) | | 主引用文献 | Elucidating Sequence and Structural Determinants of Carbohydrate Esterases for Complete Deacetylation of Substituted Xylans.
Molecules, 27, 2022
|
|
7TOI
 
 | | Crystal structure of carbohydrate esterase PbeAcXE, in complex with acetate | | 分子名称: | ACETATE ION, SGNH hydrolase | | 著者 | Stogios, P.J, Skarina, T, Di Leo, R, Jurak, E, Master, E. | | 登録日 | 2022-01-24 | | 公開日 | 2022-04-13 | | 最終更新日 | 2022-11-02 | | 実験手法 | X-RAY DIFFRACTION (1.13 Å) | | 主引用文献 | Elucidating Sequence and Structural Determinants of Carbohydrate Esterases for Complete Deacetylation of Substituted Xylans.
Molecules, 27, 2022
|
|
7TOJ
 
 | | Crystal structure of carbohydrate esterase CspAcXE, apoenzyme | | 分子名称: | CHLORIDE ION, SGNH/GDSL hydrolase family protein | | 著者 | Stogios, P.J, Skarina, T, Di Leo, R, Jurak, E, Master, E. | | 登録日 | 2022-01-24 | | 公開日 | 2022-04-13 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (1.3 Å) | | 主引用文献 | Elucidating Sequence and Structural Determinants of Carbohydrate Esterases for Complete Deacetylation of Substituted Xylans.
Molecules, 27, 2022
|
|
7TOH
 
 | | Crystal structure of carbohydrate esterase PbeAcXE, in complex with MeGlcpA-Xylp | | 分子名称: | 4-O-methyl-alpha-D-glucopyranuronic acid-(1-2)-beta-D-xylopyranose, SGNH hydrolase | | 著者 | Stogios, P.J, Skarina, T, Di Leo, R, Jurak, E, Master, E. | | 登録日 | 2022-01-24 | | 公開日 | 2022-04-13 | | 最終更新日 | 2022-11-02 | | 実験手法 | X-RAY DIFFRACTION (1.26 Å) | | 主引用文献 | Elucidating Sequence and Structural Determinants of Carbohydrate Esterases for Complete Deacetylation of Substituted Xylans.
Molecules, 27, 2022
|
|
4MUT
 
 | | Crystal structure of vancomycin resistance D,D-dipeptidase/D,D-pentapeptidase VanXYc D59S mutant in complex with D-Alanine | | 分子名称: | 3,6,9,12,15,18,21,24,27,30,33,36,39-TRIDECAOXAHENTETRACONTANE-1,41-DIOL, CHLORIDE ION, D,D-dipeptidase/D,D-carboxypeptidase, ... | | 著者 | Stogios, P.J, Evdokimova, E, Meziane-Cherif, D, Di Leo, R, Yim, V, Courvalin, P, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2013-09-23 | | 公開日 | 2013-10-09 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.25 Å) | | 主引用文献 | Structural basis for the evolution of vancomycin resistance D,D-peptidases.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
7JM1
 
 | | Crystal structure of aminoglycoside resistance enzyme ApmA, complex with acetyl-CoA | | 分子名称: | ACETYL COENZYME *A, Aminocyclitol acetyltransferase ApmA | | 著者 | Stogios, P.J, Evdokimova, E, Di Leo, R, Bordeleau, E, Wright, G.D, Savchenko, A, Joachimiak, A, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2020-07-30 | | 公開日 | 2020-09-16 | | 実験手法 | X-RAY DIFFRACTION (2.31 Å) | | 主引用文献 | Crystal structure of aminoglycoside resistance enzyme ApmA, complex with acetyl-CoA
To Be Published
|
|
7KAG
 
 | | Crystal structure of the ubiquitin-like domain 1 (Ubl1) of Nsp3 from SARS-CoV-2 | | 分子名称: | 1,2-ETHANEDIOL, Non-structural protein 3, SULFATE ION | | 著者 | Stogios, P.J, Skarina, T, Chang, C, Kim, Y, Di Leo, R, Savchenko, A, Joachimiak, A, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2020-09-30 | | 公開日 | 2020-10-14 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (3.21 Å) | | 主引用文献 | Crystal structure of the ubiquitin-like domain 1 (Ubl1) of Nsp3 from SARS-CoV-2
To Be Published
|
|
7JM2
 
 | | Crystal structure of aminoglycoside resistance enzyme ApmA, complex with apramycin | | 分子名称: | APRAMYCIN, Aminocyclitol acetyltransferase ApmA, CHLORIDE ION | | 著者 | Stogios, P.J, Evdokimova, E, Di Leo, R, Bordeleau, E, Wright, G.D, Savchenko, A, Joachimiak, A, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2020-07-30 | | 公開日 | 2020-09-16 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Crystal structure of aminoglycoside resistance enzyme ApmA, complex with apramycin
To Be Published
|
|
7JM0
 
 | | Crystal structure of aminoglycoside resistance enzyme ApmA, apoenzyme | | 分子名称: | Aminocyclitol acetyltransferase ApmA, SULFATE ION | | 著者 | Stogios, P.J, Evdokimova, E, Di Leo, R, Bordeleau, E, Wright, G.D, Savchenko, A, Joachimiak, A, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2020-07-30 | | 公開日 | 2020-09-16 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.08 Å) | | 主引用文献 | Crystal structure of aminoglycoside resistance enzyme ApmA, apoenzyme
To Be Published
|
|
4MUS
 
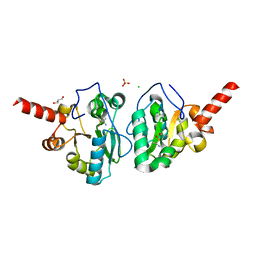 | | Crystal structure of vancomycin resistance D,D-dipeptidase/D,D-pentapeptidase VanXYc D59S mutant in complex with D-Ala-D-Ala phosphinate analog | | 分子名称: | (2R)-3-[(R)-[(1R)-1-aminoethyl](hydroxy)phosphoryl]-2-methylpropanoic acid, (2R)-3-[(R)-[(1S)-1-aminoethyl](hydroxy)phosphoryl]-2-methylpropanoic acid, CHLORIDE ION, ... | | 著者 | Stogios, P.J, Evdokimova, E, Meziane-Cherif, D, Di Leo, R, Yim, V, Courvalin, P, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2013-09-23 | | 公開日 | 2013-10-09 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.675 Å) | | 主引用文献 | Structural basis for the evolution of vancomycin resistance D,D-peptidases.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4MUR
 
 | | Crystal structure of vancomycin resistance D,D-dipeptidase/D,D-pentapeptidase VanXYc D59S mutant | | 分子名称: | 3,6,9,12,15,18,21,24,27,30,33,36,39-TRIDECAOXAHENTETRACONTANE-1,41-DIOL, CHLORIDE ION, D,D-dipeptidase/D,D-carboxypeptidase, ... | | 著者 | Stogios, P.J, Evdokimova, E, Meziane-Cherif, D, Di Leo, R, Yim, V, Courvalin, P, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2013-09-23 | | 公開日 | 2013-10-02 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | Structural basis for the evolution of vancomycin resistance D,D-peptidases.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4MUQ
 
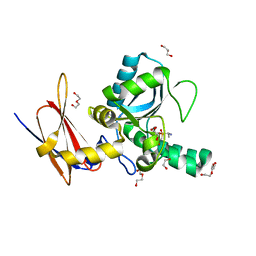 | | Crystal Structure of Vancomycin Resistance D,D-dipeptidase VanXYg in complex with D-Ala-D-Ala phosphinate analog | | 分子名称: | (2R)-3-[(R)-[(1R)-1-aminoethyl](hydroxy)phosphoryl]-2-methylpropanoic acid, (2R)-3-[(R)-[(1S)-1-aminoethyl](hydroxy)phosphoryl]-2-methylpropanoic acid, 1,2-ETHANEDIOL, ... | | 著者 | Stogios, P.J, Evdokimova, E, Meziane-Cherif, D, Di Leo, R, Yim, V, Courvalin, P, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2013-09-23 | | 公開日 | 2013-10-09 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.364 Å) | | 主引用文献 | Structural basis for the evolution of vancomycin resistance D,D-peptidases.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
