3K00
 
 | | Crystal structures of the GacH receptor of Streptomyces glaucescens GLA.O in the unliganded form and in complex with acarbose and an acarbose homolog. Comparison with acarbose-loaded maltose binding protein of Salmonella typhimurium. | | 分子名称: | Acarbose/maltose binding protein GacH, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | 著者 | Vahedi-Faridi, A, Licht, A, Bulut, H, Schneider, E. | | 登録日 | 2009-09-24 | | 公開日 | 2010-02-16 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (1.55 Å) | | 主引用文献 | Crystal Structures of the Solute Receptor GacH of Streptomyces glaucescens in Complex with Acarbose and an Acarbose Homolog: Comparison with the Acarbose-Loaded Maltose-Binding Protein of Salmonella typhimurium.
J.Mol.Biol., 397, 2010
|
|
2WEH
 
 | | Thermodynamic Optimisation of Carbonic Anhydrase Fragment Inhibitors | | 分子名称: | 2-CHLOROBENZENESULFONAMIDE, CARBONIC ANHYDRASE 2, GLYCEROL, ... | | 著者 | Scott, A.D, Phillips, C, Alex, A, Bent, A, O'Brien, R, Damian, L, Jones, L.H. | | 登録日 | 2009-03-31 | | 公開日 | 2009-11-10 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.09 Å) | | 主引用文献 | Thermodynamic Optimisation in Drug Discovery: A Case Study Using Carbonic Anhydrase Inhibitors.
Chemmedchem, 4, 2009
|
|
5DDJ
 
 | | Crystal structure of recombinant foot-and-mouth-disease virus O1M-S2093Y empty capsid | | 分子名称: | Foot and mouth disease virus, VP1, VP2, ... | | 著者 | Kotecha, A, Seago, J, Scott, K, Burman, A, Loureiro, S, Ren, J, Porta, C, Ginn, H.M, Jackson, T, Perez-Martin, E, Siebert, C.A, Paul, G, Huiskonen, J.T, Jones, I.M, Esnouf, R.M, Fry, E.E, Maree, F.F, Charleston, B, Stuart, D.I. | | 登録日 | 2015-08-25 | | 公開日 | 2015-09-23 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (3.5 Å) | | 主引用文献 | Structure-based energetics of protein interfaces guides foot-and-mouth disease virus vaccine design.
Nat.Struct.Mol.Biol., 22, 2015
|
|
1VBP
 
 | | Crystal structure of artocarpin-mannopentose complex | | 分子名称: | SULFATE ION, alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]alpha-D-mannopyranose, alpha-D-mannopyranose-(1-3)-alpha-D-mannopyranose-(1-6)-[alpha-D-mannopyranose-(1-3)]alpha-D-mannopyranose, ... | | 著者 | Jeyaprakash, A.A, Srivastav, A, Surolia, A, Vijayan, M. | | 登録日 | 2004-02-28 | | 公開日 | 2004-06-15 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (3.5 Å) | | 主引用文献 | Structural basis for the carbohydrate specificities of artocarpin: variation in the length of a loop as a strategy for generating ligand specificity
J.Mol.Biol., 338, 2004
|
|
2K4C
 
 | | tRNAPhe-based homology model for tRNAVal refined against base N-H RDCs in two media and SAXS data | | 分子名称: | 76-MER | | 著者 | Grishaev, A, Ying, J, Canny, M.D, Pardi, A, Bax, A. | | 登録日 | 2008-06-04 | | 公開日 | 2008-12-09 | | 最終更新日 | 2024-05-01 | | 実験手法 | SOLUTION NMR, SOLUTION SCATTERING | | 主引用文献 | Solution structure of tRNAVal from refinement of homology model against residual dipolar coupling and SAXS data.
J.Biomol.Nmr, 42, 2008
|
|
6VFH
 
 | |
4PP0
 
 | | Structure of the PBP NocT-M117N in complex with pyronopaline | | 分子名称: | 1,2-ETHANEDIOL, 1-[(1S)-4-carbamimidamido-1-carboxybutyl]-5-oxo-D-proline, DI(HYDROXYETHYL)ETHER, ... | | 著者 | Morera, S, Vigouroux, A. | | 登録日 | 2014-02-26 | | 公開日 | 2014-10-22 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.57 Å) | | 主引用文献 | Agrobacterium uses a unique ligand-binding mode for trapping opines and acquiring a competitive advantage in the niche construction on plant host.
Plos Pathog., 10, 2014
|
|
3K6C
 
 | | Crystal structure of protein ne0167 from nitrosomonas europaea | | 分子名称: | Uncharacterized protein NE0167 | | 著者 | Chang, C, Evdokimova, E, Savchenko, A, Edwards, A.M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2009-10-08 | | 公開日 | 2009-10-27 | | 最終更新日 | 2017-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Crystal Structure of Protein Ne0167 from Nitrosomonas Europaea
To be Published
|
|
5F0A
 
 | | CRYSTAL STRUCTURE OF PVX_084705 WITH BOUND 1-tert-butyl-3-(3-chlorophenoxy)-1H-pyrazolo[3,4-d]pyrimidin-4-amine INHIBITOR | | 分子名称: | 1-tert-butyl-3-(3-chlorophenoxy)-1H-pyrazolo[3,4-d]pyrimidin-4-amine, cGMP-dependent protein kinase, putative | | 著者 | Walker, J.R, Wernimont, A.K, He, H, Seitova, A, Loppnau, P, Sibley, L.D, Graslund, S, Hutchinson, A, Bountra, C, Weigelt, J, Edwards, A.M, Arrowsmith, C.H, Hui, R, El Bakkouri, M, Structural Genomics Consortium (SGC) | | 登録日 | 2015-11-27 | | 公開日 | 2015-12-16 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | CRYSTAL STRUCTURE OF PVX_084705 WITH BOUND INHIBITOR
To be published
|
|
6SEC
 
 | | Cold-adapted beta-D-galactosidase from Arthrobacter sp. 32cBon complex with ONPG | | 分子名称: | 2-nitrophenyl beta-D-galactopyranoside, ACETATE ION, Beta-galactosidase, ... | | 著者 | Rutkiewicz, M, Bujacz, A, Bujacz, G. | | 登録日 | 2019-07-29 | | 公開日 | 2019-09-11 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.768 Å) | | 主引用文献 | Active Site Architecture and Reaction Mechanism Determination of Cold Adapted beta-d-galactosidase fromArthrobactersp. 32cB.
Int J Mol Sci, 20, 2019
|
|
1TR0
 
 | | Crystal Structure of a boiling stable protein SP1 | | 分子名称: | GLYCEROL, stable protein 1 | | 著者 | Almog, O, Gonzalez, A, Sofer, O, Dgany, O, Shoseyov, O. | | 登録日 | 2004-06-18 | | 公開日 | 2004-09-21 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | The structural basis of the thermostability of SP1, a novel plant (Populus tremula) boiling stable protein
J.Biol.Chem., 279, 2004
|
|
5F0K
 
 | | Structure of VPS35 N terminal region | | 分子名称: | 1,2-ETHANEDIOL, GLYCEROL, SULFATE ION, ... | | 著者 | Lucas, M, Gershlick, D, Vidaurrazaga, A, Rojas, A.L, Bonifacino, J.S, Hierro, A. | | 登録日 | 2015-11-27 | | 公開日 | 2016-12-07 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (3.074 Å) | | 主引用文献 | Structural Mechanism for Cargo Recognition by the Retromer Complex.
Cell, 167, 2016
|
|
3JD1
 
 | | Glutamate dehydrogenase in complex with NADH, closed conformation | | 分子名称: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, Glutamate dehydrogenase 1, mitochondrial | | 著者 | Borgnia, M.J, Banerjee, S, Merk, A, Matthies, D, Bartesaghi, A, Rao, P, Pierson, J, Earl, L.A, Falconieri, V, Subramaniam, S, Milne, J.L.S. | | 登録日 | 2016-03-28 | | 公開日 | 2016-04-27 | | 最終更新日 | 2024-02-21 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | Using Cryo-EM to Map Small Ligands on Dynamic Metabolic Enzymes: Studies with Glutamate Dehydrogenase.
Mol.Pharmacol., 89, 2016
|
|
6SE8
 
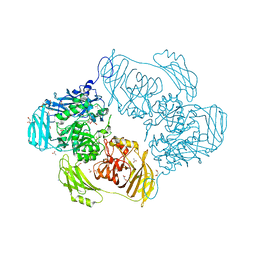 | | Cold-adapted beta-D-galactosidase from Arthrobacter sp. 32cB mutant E441Q | | 分子名称: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ACETATE ION, Beta-galactosidase, ... | | 著者 | Rutkiewicz, M, Bujacz, A, Bujacz, G. | | 登録日 | 2019-07-29 | | 公開日 | 2019-09-11 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.835 Å) | | 主引用文献 | Active Site Architecture and Reaction Mechanism Determination of Cold Adapted beta-d-galactosidase fromArthrobactersp. 32cB.
Int J Mol Sci, 20, 2019
|
|
6VFI
 
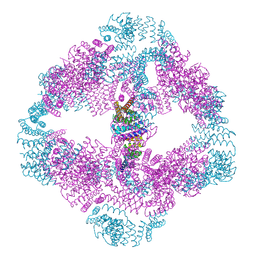 | |
6SEA
 
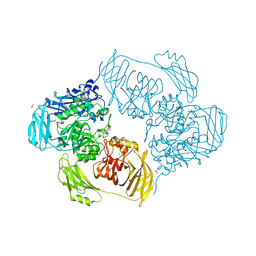 | | Cold-adapted beta-D-galactosidase from Arthrobacter sp. 32cB mutant E441Q in complex with lactose bound in deep mode | | 分子名称: | ACETATE ION, Beta-galactosidase, SODIUM ION, ... | | 著者 | Rutkiewicz, M, Bujacz, A, Bujacz, G. | | 登録日 | 2019-07-29 | | 公開日 | 2019-09-11 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.869 Å) | | 主引用文献 | Active Site Architecture and Reaction Mechanism Determination of Cold Adapted beta-d-galactosidase fromArthrobactersp. 32cB.
Int J Mol Sci, 20, 2019
|
|
6SET
 
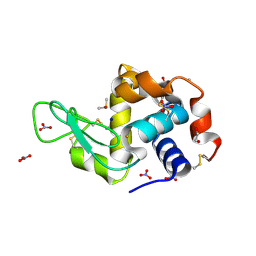 | | X-ray structure of the gold/lysozyme adduct formed upon 3 days exposure of protein crystals to compound 1 | | 分子名称: | 1,2-ETHANEDIOL, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | 著者 | Ferraro, G, Giorgio, A, Merlino, A. | | 登録日 | 2019-07-30 | | 公開日 | 2019-09-18 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Protein-mediated disproportionation of Au(i): insights from the structures of adducts of Au(iii) compounds bearing N,N-pyridylbenzimidazole derivatives with lysozyme.
Dalton Trans, 48, 2019
|
|
3ETF
 
 | | Crystal structure of a putative succinate-semialdehyde dehydrogenase from salmonella typhimurium lt2 | | 分子名称: | Putative succinate-semialdehyde dehydrogenase | | 著者 | Brunzelle, J.S, Evdokimova, E, Kudritska, M, Wawrzak, Z, Anderson, W.F, Savchenk, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2008-10-07 | | 公開日 | 2008-11-04 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Structure and activity of the NAD(P)(+) -dependent succinate semialdehyde dehydrogenase YneI from Salmonella typhimurium.
Proteins, 81, 2013
|
|
6SZ5
 
 | |
2BB3
 
 | | Crystal Structure of Cobalamin Biosynthesis Precorrin-6Y Methylase (cbiE) from Archaeoglobus fulgidus | | 分子名称: | S-ADENOSYL-L-HOMOCYSTEINE, cobalamin biosynthesis precorrin-6Y methylase (cbiE) | | 著者 | Kim, Y, Joachimiak, A, Xu, X, Savchenko, A, Edwards, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2005-10-17 | | 公開日 | 2005-11-29 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (2.27 Å) | | 主引用文献 | Crystal Structure of Cobalamin Biosynthesis Precorrin-6Y Methylase (cbiE) from Archaeoglobus fulgidus
To be Published
|
|
6SE9
 
 | | Cold-adapted beta-D-galactosidase from Arthrobacter sp. 32cB mutant E441Q in complex with lactose bound in shallow mode | | 分子名称: | ACETATE ION, Beta-galactosidase, FORMIC ACID, ... | | 著者 | Rutkiewicz, M, Bujacz, A, Bujacz, G. | | 登録日 | 2019-07-29 | | 公開日 | 2019-09-11 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.965 Å) | | 主引用文献 | Active Site Architecture and Reaction Mechanism Determination of Cold Adapted beta-d-galactosidase fromArthrobactersp. 32cB.
Int J Mol Sci, 20, 2019
|
|
2JYN
 
 | | A novel solution NMR structure of protein yst0336 from Saccharomyces cerevisiae. Northeast Structural Genomics Consortium target YT51/Ontario Centre for Structural Proteomics target yst0336 | | 分子名称: | UPF0368 protein YPL225W | | 著者 | Wu, B, Yee, A, Fares, C, Lemak, A, Gutmanas, A, Semest, A, Arrowsmith, C.H, Northeast Structural Genomics Consortium (NESG), Ontario Centre for Structural Proteomics (OCSP) | | 登録日 | 2007-12-14 | | 公開日 | 2007-12-25 | | 最終更新日 | 2024-05-29 | | 実験手法 | SOLUTION NMR | | 主引用文献 | A novel solution NMR structure of protein yst0336 from Saccharomyces cerevisiae.
To be Published
|
|
1YQE
 
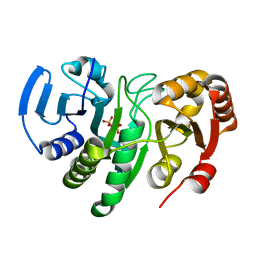 | | Crystal Structure of Conserved Protein of Unknown Function AF0625 | | 分子名称: | Hypothetical UPF0204 protein AF0625, PYROPHOSPHATE 2- | | 著者 | Liu, Y, Skarina, T, Dong, A, Kudritskam, M, Savchenko, A, Pai, E.F, Edwards, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2005-02-01 | | 公開日 | 2005-03-15 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.83 Å) | | 主引用文献 | Crystal Structure of Conserved Hypothetical Protein AF0625
To be Published
|
|
6T0J
 
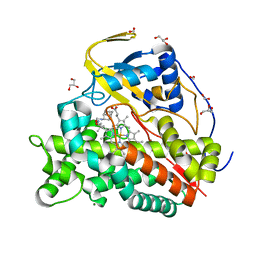 | | Crystal structure of CYP124 in complex with SQ109 | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, FORMIC ACID, GLYCEROL, ... | | 著者 | Bukhdruker, S, Marin, E, Varaksa, T, Gilep, A, Strushkevich, N, Borshchevskiy, V. | | 登録日 | 2019-10-03 | | 公開日 | 2020-10-14 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.25 Å) | | 主引用文献 | Hydroxylation of Antitubercular Drug Candidate, SQ109, by Mycobacterial Cytochrome P450.
Int J Mol Sci, 21, 2020
|
|
5EUV
 
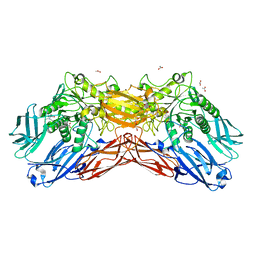 | | Crystal structure of a cold-adapted dimeric beta-D-galactosidase from Paracoccus sp. 32d strain | | 分子名称: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETATE ION, Beta-D-galactosidase, ... | | 著者 | Rutkiewicz-Krotewicz, M, Bujacz, A, Pietrzyk, A.J, Sekula, B, Bujacz, G. | | 登録日 | 2015-11-19 | | 公開日 | 2016-09-14 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Structural studies of a cold-adapted dimeric beta-D-galactosidase from Paracoccus sp. 32d.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
