7C7P
 
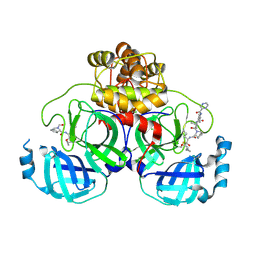 | | Crystal structure of the SARS-CoV-2 main protease in complex with Telaprevir | | 分子名称: | (1S,3aR,6aS)-2-[(2S)-2-({(2S)-2-cyclohexyl-2-[(pyrazin-2-ylcarbonyl)amino]acetyl}amino)-3,3-dimethylbutanoyl]-N-[(2R,3S)-1-(cyclopropylamino)-2-hydroxy-1-oxohexan-3-yl]octahydrocyclopenta[c]pyrrole-1-carboxamide, 3C-like proteinase, CHLORIDE ION | | 著者 | Zeng, R, Qiao, J.X, Wang, Y.F, Li, Y.S, Yao, R, Yang, S.Y, Lei, J. | | 登録日 | 2020-05-26 | | 公開日 | 2020-07-29 | | 最終更新日 | 2024-11-06 | | 実験手法 | X-RAY DIFFRACTION (1.74 Å) | | 主引用文献 | SARS-CoV-2 M pro inhibitors with antiviral activity in a transgenic mouse model.
Science, 371, 2021
|
|
7COM
 
 | | Crystal structure of the SARS-CoV-2 main protease in complex with Boceprevir (space group P212121) | | 分子名称: | 3C-like proteinase, boceprevir (bound form) | | 著者 | Zeng, R, Qiao, J.X, Wang, Y.F, Li, Y.S, Yao, R, Liu, J.M, Zhou, Y.L, Chen, P, Yang, S.Y, Lei, J. | | 登録日 | 2020-08-04 | | 公開日 | 2020-08-19 | | 最終更新日 | 2024-11-06 | | 実験手法 | X-RAY DIFFRACTION (2.25 Å) | | 主引用文献 | SARS-CoV-2 M pro inhibitors with antiviral activity in a transgenic mouse model.
Science, 371, 2021
|
|
6A73
 
 | | Complex structure of CSN2 with IP6 | | 分子名称: | COP9 signalosome complex subunit 2,Endolysin, INOSITOL HEXAKISPHOSPHATE, SULFATE ION | | 著者 | Liu, L, Li, D, Rao, F, Wang, T. | | 登録日 | 2018-07-02 | | 公開日 | 2019-07-03 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.447 Å) | | 主引用文献 | Basis for metabolite-dependent Cullin-RING ligase deneddylation by the COP9 signalosome.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
3V86
 
 | |
5ZX1
 
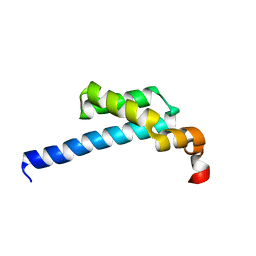 | |
4HXX
 
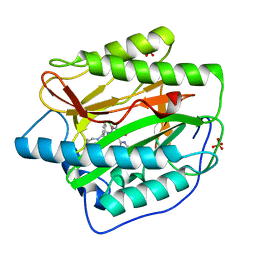 | | Pyridinylpyrimidines selectively inhibit human methionine aminopeptidase-1 | | 分子名称: | (1R)-N~2~-[5-chloro-2-(5-chloropyridin-2-yl)-6-methylpyrimidin-4-yl]-1-phenyl-N~1~-(4-phenylbutyl)ethane-1,2-diamine, COBALT (II) ION, Methionine aminopeptidase 1, ... | | 著者 | Gabelli, S.B, Zhang, F, Liu, J, Amzel, L.M. | | 登録日 | 2012-11-12 | | 公開日 | 2013-04-03 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.09 Å) | | 主引用文献 | Pyridinylpyrimidines selectively inhibit human methionine aminopeptidase-1.
Bioorg.Med.Chem., 21, 2013
|
|
1UP7
 
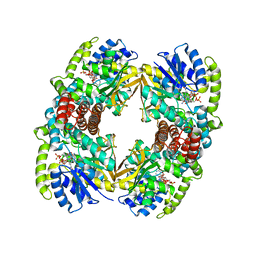 | | Structure of the 6-phospho-beta glucosidase from Thermotoga maritima at 2.4 Angstrom resolution in the tetragonal form with NAD and glucose-6-phosphate | | 分子名称: | 6-O-phosphono-alpha-D-glucopyranose, 6-PHOSPHO-BETA-GLUCOSIDASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | 著者 | Varrot, A, Yip, V.L, Withers, S.G, Davies, G.J. | | 登録日 | 2003-09-29 | | 公開日 | 2004-11-18 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Nad+ and Metal-Ion Dependent Hydrolysis by Family 4 Glycosidases: Structural Insight Into Specificity for Phospho-Beta-D-Glucosides
J.Mol.Biol., 346, 2005
|
|
1UP6
 
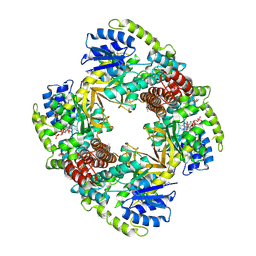 | | Structure of the 6-phospho-beta glucosidase from Thermotoga maritima at 2.55 Angstrom resolution in the tetragonal form with manganese, NAD+ and glucose-6-phosphate | | 分子名称: | 6-O-phosphono-alpha-D-glucopyranose, 6-PHOSPHO-BETA-GLUCOSIDASE, MANGANESE (II) ION, ... | | 著者 | Varrot, A, Yip, V.L, Withers, S.G, Davies, G.J. | | 登録日 | 2003-09-29 | | 公開日 | 2004-08-02 | | 最終更新日 | 2024-10-09 | | 実験手法 | X-RAY DIFFRACTION (2.55 Å) | | 主引用文献 | An Unusual Mechanism of Glycoside Hydrolysis Involving Redox and Elimination Steps by a Family 4 Beta-Glycosidase from Thermotoga Maritima.
J.Am.Chem.Soc., 126, 2004
|
|
1UP4
 
 | |
5G06
 
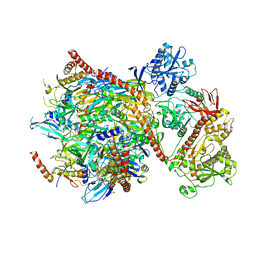 | | Cryo-EM structure of yeast cytoplasmic exosome | | 分子名称: | EXOSOME COMPLEX COMPONENT CSL4, EXOSOME COMPLEX COMPONENT MTR3, EXOSOME COMPLEX COMPONENT RRP4, ... | | 著者 | Liu, J.J, Niu, C.Y, Wu, Y, Tan, D, Wang, Y, Ye, M.D, Liu, Y, Zhao, W.W, Zhou, K, Liu, Q.S, Dai, J.B, Yang, X.R, Dong, M.Q, Huang, N, Wang, H.W. | | 登録日 | 2016-03-17 | | 公開日 | 2016-06-15 | | 最終更新日 | 2024-11-06 | | 実験手法 | ELECTRON MICROSCOPY (4.2 Å) | | 主引用文献 | Cryoem Structure of Yeast Cytoplasmic Exosome Complex.
Cell Res., 26, 2016
|
|
7D3I
 
 | | Crystal structure of SARS-CoV-2 main protease in complex with MI-23 | | 分子名称: | (3~{S},3~{a}~{S},6~{a}~{R})-2-[3-[3,5-bis(fluoranyl)phenyl]propanoyl]-~{N}-[(2~{S})-1-oxidanylidene-3-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]propan-2-yl]-3,3~{a},4,5,6,6~{a}-hexahydro-1~{H}-cyclopenta[c]pyrrole-3-carboxamide, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3C-like proteinase | | 著者 | Zeng, R, Li, Y.S, Qiao, J.X, Wang, Y.F, Yang, S.Y, Lei, J. | | 登録日 | 2020-09-19 | | 公開日 | 2020-10-07 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.004 Å) | | 主引用文献 | SARS-CoV-2 M pro inhibitors with antiviral activity in a transgenic mouse model.
Science, 371, 2021
|
|
5K9I
 
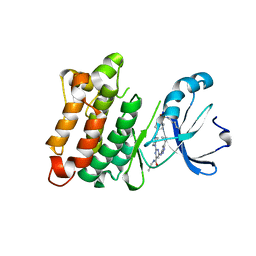 | | Crystal structure of c-SRC in complex with a covalent lysine probe | | 分子名称: | 4-[(4-{4-[(3-cyclopropyl-1H-pyrazol-5-yl)amino]-6-[(prop-2-yn-1-yl)carbamoyl]pyrimidin-2-yl}piperazin-1-yl)methyl]benzene-1-sulfonyl fluoride, Proto-oncogene tyrosine-protein kinase Src | | 著者 | Wan, X, Ouyang, S, Zhao, Q, Taunton, J. | | 登録日 | 2016-05-31 | | 公開日 | 2017-02-01 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Broad-Spectrum Kinase Profiling in Live Cells with Lysine-Targeted Sulfonyl Fluoride Probes.
J. Am. Chem. Soc., 139, 2017
|
|
7L28
 
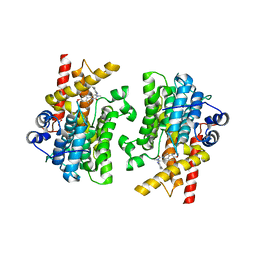 | | Crystal structure of the catalytic domain of human PDE3A bound to Trequinsin | | 分子名称: | (2E)-9,10-dimethoxy-3-methyl-2-[(2,4,6-trimethylphenyl)imino]-2,3,6,7-tetrahydro-4H-pyrimido[6,1-a]isoquinolin-4-one, ACETATE ION, MAGNESIUM ION, ... | | 著者 | Horner, S.W, Garvie, C. | | 登録日 | 2020-12-16 | | 公開日 | 2021-06-16 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Structure of PDE3A-SLFN12 complex reveals requirements for activation of SLFN12 RNase.
Nat Commun, 12, 2021
|
|
7L29
 
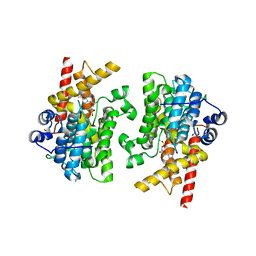 | |
7L27
 
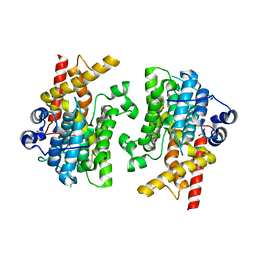 | |
7KWE
 
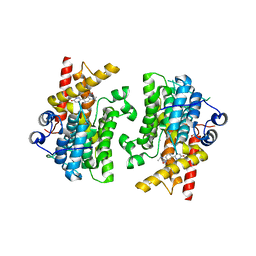 | |
2HE2
 
 | | Crystal structure of the 3rd PDZ domain of human discs large homologue 2, DLG2 | | 分子名称: | Discs large homolog 2 | | 著者 | Turnbull, A.P, Phillips, C, Berridge, G, Savitsky, P, Smee, C.E.A, Papagrigoriou, E, Debreczeni, J, Gorrec, F, Elkins, J.M, von Delft, F, Weigelt, J, Edwards, A, Arrowsmith, C, Sundstrom, M, Doyle, D.A, Structural Genomics Consortium (SGC) | | 登録日 | 2006-06-21 | | 公開日 | 2006-07-04 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Structure of PICK1 and other PDZ domains obtained with the help of self-binding C-terminal extensions.
Protein Sci., 16, 2007
|
|
8IPT
 
 | |
8IPS
 
 | |
8IPR
 
 | |
8IPQ
 
 | |
2ZGJ
 
 | | Crystal Structure of D86N-GzmM Complexed with Its Optimal Synthesized Substrate | | 分子名称: | Granzyme M, SSGKVPLS, SULFATE ION | | 著者 | Wu, L.F, Wang, L, Hua, G.Q, Liu, K, Zhai, Y.J, Sun, F, Fan, Z.S. | | 登録日 | 2008-01-22 | | 公開日 | 2009-01-27 | | 最終更新日 | 2024-10-30 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Structural basis for proteolytic specificity of the human apoptosis-inducing granzyme M
J.Immunol., 183, 2009
|
|
2ZGH
 
 | | Crystal Structure of active granzyme M bound to its product | | 分子名称: | Granzyme M, SSGKVPL, SULFATE ION | | 著者 | Wu, L.F, Wang, L, Hua, G.Q, Liu, K, Zhai, Y.J, Sun, F, Fan, Z.S. | | 登録日 | 2008-01-22 | | 公開日 | 2009-01-27 | | 最終更新日 | 2024-11-13 | | 実験手法 | X-RAY DIFFRACTION (2.17 Å) | | 主引用文献 | Structural basis for proteolytic specificity of the human apoptosis-inducing granzyme M
J.Immunol., 183, 2009
|
|
2ICA
 
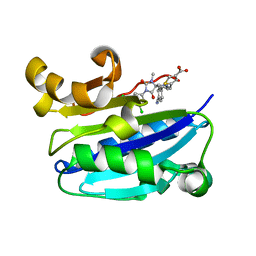 | | CD11a (LFA1) I-domain complexed with BMS-587101 aka 5-[(5S, 9R)-9-(4-cyanophenyl)-3-(3,5-dichlorophenyl)-1-methyl-2,4-dioxo-1,3,7-triazaspiro [4.4]non-7-yl]methyl]-3-thiophenecarboxylicacid | | 分子名称: | 5-[(5S,9R)-9-(4-CYANOPHENYL)-3-(3,5-DICHLOROPHENYL)-1-METHYL-2,4-DIOXO-1,3,7-TRIAZASPIRO [4.4]NON-7-YL]METHYL]-3-THIOPHENECARBOXYLICACID, Integrin alpha-L | | 著者 | Sheriff, S, Einspahr, H. | | 登録日 | 2006-09-12 | | 公開日 | 2006-12-19 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (1.56 Å) | | 主引用文献 | Discovery and Development of 5-[(5S,9R)-9- (4-Cyanophenyl)-3-(3,5-dichlorophenyl)-1- methyl-2,4-dioxo-1,3,7-triazaspiro[4.4]non- 7-yl-methyl]-3-thiophenecarboxylic acid (BMS-587101)-A Small Molecule Antagonist Leukocyte Function Associated Antigen-1.
J.Med.Chem., 49, 2006
|
|
6LLG
 
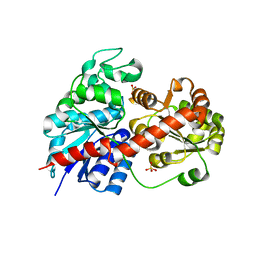 | | Crystal Structure of Fagopyrum esculentum M UGT708C1 | | 分子名称: | BENZAMIDINE, SULFATE ION, UDP-glycosyltransferase 708C1 | | 著者 | Wang, X, Liu, M. | | 登録日 | 2019-12-23 | | 公開日 | 2020-09-09 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Crystal Structures of theC-Glycosyltransferase UGT708C1 from Buckwheat Provide Insights into the Mechanism ofC-Glycosylation.
Plant Cell, 32, 2020
|
|
