7V8Z
 
 | |
8OST
 
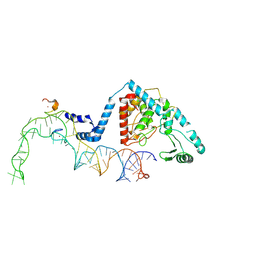 | | Structure of human terminal uridylyltransferase 4 (TUT4, ZCCHC11) in complex with pre-let7g miRNA and Lin28A | | 分子名称: | Protein lin-28 homolog A, Terminal uridylyltransferase 4, ZINC ION, ... | | 著者 | Gilbert, R.J, Yi, G, Ye, M. | | 登録日 | 2023-04-20 | | 公開日 | 2024-07-17 | | 最終更新日 | 2024-07-24 | | 実験手法 | ELECTRON MICROSCOPY (3.69 Å) | | 主引用文献 | Structural basis for activity switching in polymerases determining the fate of let-7 pre-miRNAs
Nat.Struct.Mol.Biol., 2024
|
|
6DUF
 
 | | Crystal structure of HIV-1 reverse transcriptase V106A/F227L mutant in complex with non-nucleoside inhibitor 25a | | 分子名称: | 1,2-ETHANEDIOL, 4-({4-[(4-{4-[(E)-2-cyanoethenyl]-2,6-dimethylphenoxy}thieno[3,2-d]pyrimidin-2-yl)amino]piperidin-1-yl}methyl)benzene-1-sulfonamide, DIMETHYL SULFOXIDE, ... | | 著者 | Yang, Y, Nguyen, L.A, Smithline, Z.B, Steitz, T.A. | | 登録日 | 2018-06-20 | | 公開日 | 2018-08-01 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.963 Å) | | 主引用文献 | Structural basis for potent and broad inhibition of HIV-1 RT by thiophene[3,2-d]pyrimidine non-nucleoside inhibitors.
Elife, 7, 2018
|
|
6DUH
 
 | | Crystal structure of HIV-1 reverse transcriptase Y181I mutant in complex with non-nucleoside inhibitor 25a | | 分子名称: | 1,2-ETHANEDIOL, 4-({4-[(4-{4-[(E)-2-cyanoethenyl]-2,6-dimethylphenoxy}thieno[3,2-d]pyrimidin-2-yl)amino]piperidin-1-yl}methyl)benzene-1-sulfonamide, DIMETHYL SULFOXIDE, ... | | 著者 | Yang, Y, Nguyen, L.A, Smithline, Z.B, Steitz, T.A. | | 登録日 | 2018-06-20 | | 公開日 | 2018-08-01 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.003 Å) | | 主引用文献 | Structural basis for potent and broad inhibition of HIV-1 RT by thiophene[3,2-d]pyrimidine non-nucleoside inhibitors.
Elife, 7, 2018
|
|
5MSL
 
 | |
5APH
 
 | | Ligand complex of RORg LBD | | 分子名称: | DIMETHYL SULFOXIDE, N-(2-FLUOROPHENYL)-4-[(4-FLUOROPHENYL)SULFONYL]-2,3,4,5-TETRAHYDRO-1,4-BENZOXAZEPIN-6-AMINE, NUCLEAR RECEPTOR COACTIVATOR 2, ... | | 著者 | Xue, Y, Aagaard, A, Narjes, F. | | 登録日 | 2015-09-16 | | 公開日 | 2015-11-25 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.54 Å) | | 主引用文献 | Benzoxazepines Achieve Potent Suppression of IL-17 Release in Human T-Helper 17 (TH 17) Cells through an Induced-Fit Binding Mode to the Nuclear Receptor ROR gamma.
ChemMedChem, 11, 2016
|
|
7VIL
 
 | |
1DK6
 
 | | NMR structure analysis of the DNA nine base pair duplex D(CATGAGTAC) D(GTAC(NP3)CATG) | | 分子名称: | 5'-D(CP*AP*TP*GP*AP*GP*TP*AP*CP*)-3', 5'-D(GP*TP*AP*CP*(NP3)P*CP*AP*TP*GP*)-3' | | 著者 | Klewer, D.A, Hoskins, A, Davisson, V.J, Bergstrom, D.E, LiWang, A.C. | | 登録日 | 1999-12-06 | | 公開日 | 2000-01-11 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | NMR structure of a DNA duplex containing nucleoside analog 1-(2'-deoxy-beta-D-ribofuranosyl)-3-nitropyrrole and the structure of the unmodified control.
Nucleic Acids Res., 28, 2000
|
|
6I1S
 
 | | Crystal structure of the ACVR1 (ALK2) kinase in complex with FKBP12 and the inhibitor E6201 | | 分子名称: | (4~{S},5~{R},6~{Z},9~{S},10~{S},12~{E})-16-(ethylamino)-4,5-dimethyl-9,10,18-tris(oxidanyl)-3-oxabicyclo[12.4.0]octadeca-1(14),6,12,15,17-pentaene-2,8-dione, 1,2-ETHANEDIOL, Activin receptor type-1, ... | | 著者 | Williams, E.P, Pinkas, D.M, Fortin, J, Newman, J.A, Bradshaw, W.J, Mahajan, P, Kupinska, K, Burgess-Brown, N, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Bullock, A.N. | | 登録日 | 2018-10-30 | | 公開日 | 2019-09-11 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.52 Å) | | 主引用文献 | Mutant ACVR1 Arrests Glial Cell Differentiation to Drive Tumorigenesis in Pediatric Gliomas.
Cancer Cell, 37, 2020
|
|
5APJ
 
 | | Ligand complex of RORg LBD | | 分子名称: | 2-CHLORO-6-FLUORO-N-[4-[3-(TRIFLUOROMETHYL)PHENYL]SULFONYL-3,5-DIHYDRO-2H-1,4-BENZOXAZEPIN-7-YL]BENZAMIDE, NUCLEAR RECEPTOR COACTIVATOR 2, NUCLEAR RECEPTOR ROR-GAMMA, ... | | 著者 | Xue, Y, Aagaard, A, Narjes, F. | | 登録日 | 2015-09-16 | | 公開日 | 2015-11-25 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.08 Å) | | 主引用文献 | Benzoxazepines Achieve Potent Suppression of IL-17 Release in Human T-Helper 17 (TH 17) Cells through an Induced-Fit Binding Mode to the Nuclear Receptor ROR gamma.
ChemMedChem, 11, 2016
|
|
5YE4
 
 | | Crystal structure of the complex of di-acetylated histone H4 and 1A9D7 Fab fragment | | 分子名称: | 1A9D7 L chain, 1A9D7 VH CH1 chain, ZINC ION, ... | | 著者 | Matsuda, T, Ito, T, Wakamori, M, Umehara, T. | | 登録日 | 2017-09-15 | | 公開日 | 2018-08-22 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.799 Å) | | 主引用文献 | JQ1 affects BRD2-dependent and independent transcription regulation without disrupting H4-hyperacetylated chromatin states.
Epigenetics, 13, 2018
|
|
7FHR
 
 | | Crystal Structure of a Rieske Oxygenase from Cupriavidus metallidurans | | 分子名称: | 1,2-ETHANEDIOL, FE (II) ION, FE2/S2 (INORGANIC) CLUSTER, ... | | 著者 | Mahto, J.K, Dhankhar, P, Kumar, P. | | 登録日 | 2021-07-30 | | 公開日 | 2021-12-15 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (1.84 Å) | | 主引用文献 | Molecular insights into substrate recognition and catalysis by phthalate dioxygenase from Comamonas testosteroni.
J.Biol.Chem., 297, 2021
|
|
3NFG
 
 | |
1DK9
 
 | | SOLUTION STRUCTURE ANALYSIS OF THE DNA DUPLEX D(CATGAGTAC)D(GTACTCATG) | | 分子名称: | 5'-D(CP*AP*TP*GP*AP*GP*TP*AP*CP*)-3', 5'-D(GP*TP*AP*CP*TP*CP*AP*TP*GP*)-3' | | 著者 | Klewer, D.A, Hoskins, A, Davisson, V.J, Bergstrom, D.E, LiWang, A.C. | | 登録日 | 1999-12-06 | | 公開日 | 1999-12-16 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | NMR structure of a DNA duplex containing nucleoside analog 1-(2'-deoxy-beta-D-ribofuranosyl)-3-nitropyrrole and the structure of the unmodified control.
Nucleic Acids Res., 28, 2000
|
|
3RS1
 
 | | Mouse C-type lectin-related protein Clrg | | 分子名称: | C-type lectin domain family 2 member I, CHLORIDE ION | | 著者 | Skalova, T, Dohnalek, J, Duskova, J, Stepankova, A, Koval, T, Hasek, J, Kotynkova, K, Vanek, O, Bezouska, K. | | 登録日 | 2011-05-02 | | 公開日 | 2012-05-02 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.94 Å) | | 主引用文献 | Mouse Clr-g, a Ligand for NK Cell Activation Receptor NKR-P1F: Crystal Structure and Biophysical Properties.
J.Immunol., 189, 2012
|
|
4HVA
 
 | |
7XML
 
 | |
7CK1
 
 | | Crystal structure of arabidopsis CESA3 catalytic domain | | 分子名称: | Cellulose synthase A catalytic subunit 3 [UDP-forming],Cellulose synthase A catalytic subunit 3 [UDP-forming], MANGANESE (II) ION | | 著者 | Qiao, Z, Gao, Y.G. | | 登録日 | 2020-07-15 | | 公開日 | 2021-03-17 | | 最終更新日 | 2021-03-31 | | 実験手法 | X-RAY DIFFRACTION (2.35 Å) | | 主引用文献 | Structure of Arabidopsis CESA3 catalytic domain with its substrate UDP-glucose provides insight into the mechanism of cellulose synthesis.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7CK3
 
 | | Crystal structure of Arabidopsis CESA3 catalytic domain | | 分子名称: | Cellulose synthase A catalytic subunit 3 [UDP-forming],Cellulose synthase A catalytic subunit 3 [UDP-forming] | | 著者 | Qiao, Z, Gao, Y.G. | | 登録日 | 2020-07-15 | | 公開日 | 2021-03-17 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Structure of Arabidopsis CESA3 catalytic domain with its substrate UDP-glucose provides insight into the mechanism of cellulose synthesis.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7CK2
 
 | | Crystal structure of Arabidopsis CESA3 catalytic domain with UDP-Glucose | | 分子名称: | Cellulose synthase A catalytic subunit 3 [UDP-forming],Cellulose synthase A catalytic subunit 3 [UDP-forming], MANGANESE (II) ION, URIDINE-5'-DIPHOSPHATE-GLUCOSE | | 著者 | Qiao, Z, Gao, Y.G. | | 登録日 | 2020-07-15 | | 公開日 | 2021-03-17 | | 最終更新日 | 2021-03-31 | | 実験手法 | X-RAY DIFFRACTION (2.05 Å) | | 主引用文献 | Structure of Arabidopsis CESA3 catalytic domain with its substrate UDP-glucose provides insight into the mechanism of cellulose synthesis.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
1CEA
 
 | |
6UL5
 
 | | Crystal structure of HIV-1 reverse transcriptase (RT) in complex with 4-[(4-{4-[(E)-2-cyanoethenyl]-2,6-dimethylphenoxy}thieno[3,2-d]pyrimidin-2-yl)amino]-2-fluorobenzonitrile (24b), a non-nucleoside RT inhibitor | | 分子名称: | 1,2-ETHANEDIOL, 4-[(4-{4-[(E)-2-cyanoethenyl]-2,6-dimethylphenoxy}thieno[3,2-d]pyrimidin-2-yl)amino]-2-fluorobenzonitrile, MAGNESIUM ION, ... | | 著者 | Ruiz, F.X, Pilch, A, Arnold, E. | | 登録日 | 2019-10-06 | | 公開日 | 2020-02-05 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.23 Å) | | 主引用文献 | Discovery and Characterization of Fluorine-Substituted Diarylpyrimidine Derivatives as Novel HIV-1 NNRTIs with Highly Improved Resistance Profiles and Low Activity for the hERG Ion Channel.
J.Med.Chem., 63, 2020
|
|
6B2I
 
 | |
6B2G
 
 | |
6B2H
 
 | |
