7RZR
 
 | |
7RZT
 
 | |
7RZV
 
 | |
8J1X
 
 | |
8J1W
 
 | |
7E7E
 
 | | The co-crystal structure of ACE2 with Fab | | 分子名称: | Processed angiotensin-converting enzyme 2, ZINC ION, h11B11-Fab | | 著者 | Xiao, J.Y, Zhang, Y. | | 登録日 | 2021-02-26 | | 公開日 | 2021-07-14 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (3.8 Å) | | 主引用文献 | A broadly neutralizing humanized ACE2-targeting antibody against SARS-CoV-2 variants.
Nat Commun, 12, 2021
|
|
6VWU
 
 | |
3CAW
 
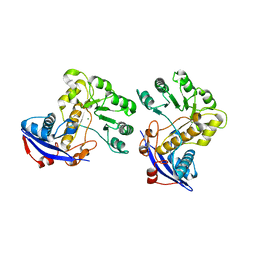 | | Crystal structure of o-succinylbenzoate synthase from Bdellovibrio bacteriovorus liganded with Mg | | 分子名称: | MAGNESIUM ION, o-succinylbenzoate synthase | | 著者 | Fedorov, A.A, Fedorov, E.V, Sakai, A, Burley, S.K, Gerlt, J.A, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | 登録日 | 2008-02-20 | | 公開日 | 2008-03-04 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (1.87 Å) | | 主引用文献 | Loss of quaternary structure is associated with rapid sequence divergence in the OSBS family.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
7END
 
 | |
7EN9
 
 | | Crystal structure of SARS-CoV-2 3CLpro in complex with the non-covalent inhibitor WU-02 | | 分子名称: | 3C-like proteinase, 5-bromanyl-~{N}-methyl-3-nitro-2-[(4~{R},5~{S})-2-(7-oxidanylisoquinolin-4-yl)carbonyl-4-phenyl-2,7-diazaspiro[4.4]nonan-7-yl]benzamide | | 著者 | Hou, N, Peng, C, Hu, Q. | | 登録日 | 2021-04-16 | | 公開日 | 2022-07-20 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Development of Highly Potent Noncovalent Inhibitors of SARS-CoV-2 3CLpro.
Acs Cent.Sci., 9, 2023
|
|
7EN8
 
 | | Crystal structure of SARS-CoV-2 3CLpro in complex with the non-covalent inhibitor WU-04 | | 分子名称: | 3C-like proteinase, GLYCEROL, ~{N}-[(1~{S},2~{R})-2-[[4-bromanyl-2-(methylcarbamoyl)-6-nitro-phenyl]amino]cyclohexyl]isoquinoline-4-carboxamide | | 著者 | Hou, N, Peng, C, Hu, Q. | | 登録日 | 2021-04-16 | | 公開日 | 2022-07-20 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.83 Å) | | 主引用文献 | Development of Highly Potent Noncovalent Inhibitors of SARS-CoV-2 3CLpro.
Acs Cent.Sci., 9, 2023
|
|
7ENE
 
 | |
8WHA
 
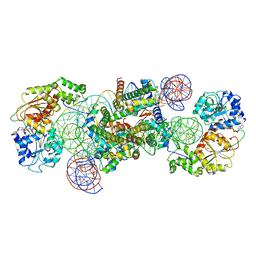 | | Structure of DDM1-nucleosome complex in the ADP-BeFx state with DDM1 bound to SHL2 and SHL-2 | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent DNA helicase DDM1, BERYLLIUM TRIFLUORIDE ION, ... | | 著者 | Liu, Y, Zhang, Z, Du, J. | | 登録日 | 2023-09-22 | | 公開日 | 2024-04-17 | | 実験手法 | ELECTRON MICROSCOPY (4.05 Å) | | 主引用文献 | Molecular basis of chromatin remodelling by DDM1 involved in plant DNA methylation.
Nat.Plants, 10, 2024
|
|
8WH5
 
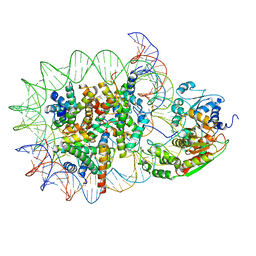 | | Structure of DDM1-nucleosome complex in the apo state | | 分子名称: | ATP-dependent DNA helicase DDM1, DNA (antisense strand), DNA (sense strand), ... | | 著者 | Liu, Y, Zhang, Z, Du, J. | | 登録日 | 2023-09-22 | | 公開日 | 2024-04-17 | | 実験手法 | ELECTRON MICROSCOPY (3.58 Å) | | 主引用文献 | Molecular basis of chromatin remodelling by DDM1 involved in plant DNA methylation.
Nat.Plants, 10, 2024
|
|
8WHB
 
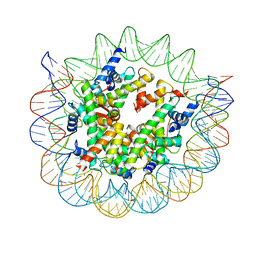 | | Structure of nucleosome core particle of Arabidopsis thaliana | | 分子名称: | DNA (antisense strand), DNA (sense strand), Histone H2A.6, ... | | 著者 | Liu, Y, Zhang, Z, Du, J. | | 登録日 | 2023-09-23 | | 公開日 | 2024-04-17 | | 実験手法 | ELECTRON MICROSCOPY (3.17 Å) | | 主引用文献 | Molecular basis of chromatin remodelling by DDM1 involved in plant DNA methylation.
Nat.Plants, 10, 2024
|
|
8WH8
 
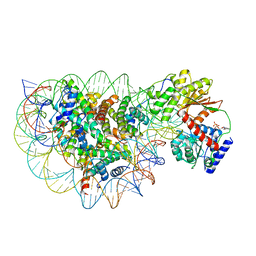 | | Structure of DDM1-nucleosome complex in ADP state | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent DNA helicase DDM1, DNA (antisense strand), ... | | 著者 | Liu, Y, Zhang, Z, Du, J. | | 登録日 | 2023-09-22 | | 公開日 | 2024-04-17 | | 実験手法 | ELECTRON MICROSCOPY (3.6 Å) | | 主引用文献 | Molecular basis of chromatin remodelling by DDM1 involved in plant DNA methylation.
Nat.Plants, 10, 2024
|
|
8WH9
 
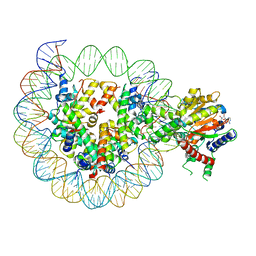 | | Structure of DDM1-nucleosome complex in ADP-BeFx state | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent DNA helicase DDM1, BERYLLIUM TRIFLUORIDE ION, ... | | 著者 | Liu, Y, Zhang, Z, Du, J. | | 登録日 | 2023-09-22 | | 公開日 | 2024-04-17 | | 実験手法 | ELECTRON MICROSCOPY (3.31 Å) | | 主引用文献 | Molecular basis of chromatin remodelling by DDM1 involved in plant DNA methylation.
Nat.Plants, 10, 2024
|
|
6AKJ
 
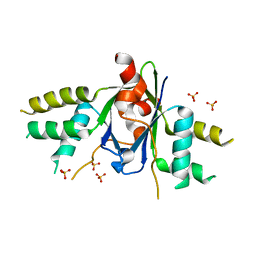 | | The crystal structure of EMC complex | | 分子名称: | Enhancer of rudimentary homolog,YTH domain-containing protein mmi1 fusion protein, SULFATE ION | | 著者 | Li, F. | | 登録日 | 2018-09-01 | | 公開日 | 2019-02-13 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | A conserved dimer interface connects ERH and YTH family proteins to promote gene silencing.
Nat Commun, 10, 2019
|
|
8Y33
 
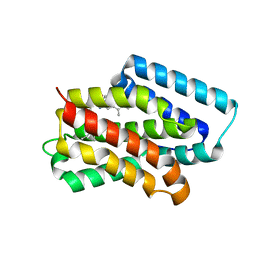 | | A near-infrared fluorescent protein of de novo backbone design | | 分子名称: | 3-[5-[(4-ethenyl-3-methyl-5-oxidanylidene-pyrrol-2-yl)methyl]-2-[[5-[(3-ethyl-4-methyl-5-oxidanylidene-pyrrol-2-yl)methyl]-3-(3-hydroxy-3-oxopropyl)-4-methyl-1~{H}-pyrrol-2-yl]methyl]-4-methyl-1~{H}-pyrrol-3-yl]propanoic acid, near-infrared fluorescent protein | | 著者 | Hu, X, Xu, Y. | | 登録日 | 2024-01-28 | | 公開日 | 2024-02-28 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Using Protein Design and Directed Evolution to Monomerize a Bright Near-Infrared Fluorescent Protein.
Acs Synth Biol, 13, 2024
|
|
5XSJ
 
 | | XylFII-LytSN complex | | 分子名称: | Periplasmic binding protein/LacI transcriptional regulator, Signal transduction histidine kinase, LytS, ... | | 著者 | Li, J.X, Wang, C.Y, Zhang, P. | | 登録日 | 2017-06-14 | | 公開日 | 2017-08-02 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (2.202 Å) | | 主引用文献 | Molecular mechanism of environmental d-xylose perception by a XylFII-LytS complex in bacteria
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5XSD
 
 | | XylFII-LytSN complex mutant - D103A | | 分子名称: | Periplasmic binding protein/LacI transcriptional regulator, Signal transduction histidine kinase, LytS | | 著者 | Li, J.X, Wang, C.Y, Zhang, P. | | 登録日 | 2017-06-13 | | 公開日 | 2017-08-02 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Molecular mechanism of environmental d-xylose perception by a XylFII-LytS complex in bacteria
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5XSS
 
 | | XylFII molecule | | 分子名称: | Periplasmic binding protein/LacI transcriptional regulator, beta-D-xylopyranose | | 著者 | Li, J.X, Wang, C.Y, Zhang, P. | | 登録日 | 2017-06-15 | | 公開日 | 2017-08-02 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (2.093 Å) | | 主引用文献 | Molecular mechanism of environmental d-xylose perception by a XylFII-LytS complex in bacteria
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
3IFJ
 
 | |
3INW
 
 | | HSP90 N-TERMINAL DOMAIN with pochoxime A | | 分子名称: | (5E,9E,11E)-13-chloro-14,16-dihydroxy-3,4,7,8-tetrahydro-1H-2-benzoxacyclotetradecine-1,11(12H)-dione 11-[O-(2-oxo-2-piperidin-1-ylethyl)oxime], DIMETHYL SULFOXIDE, Heat shock protein HSP 90-alpha | | 著者 | Korndoerfer, I.P. | | 登録日 | 2009-08-13 | | 公開日 | 2010-08-18 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Inhibition of HSP90 with pochoximes: SAR and structure-based insights.
Chembiochem, 10, 2009
|
|
7CU2
 
 | | CRYSTAL STRUCTURE OF STREPTOMYCES ALBOGRISEOLUS FLAVIN-DEPENDENT TRYPTOPHAN 6-HALOGENASE THAL IN COMPLEX WITH REDUCED FAD | | 分子名称: | DIHYDROFLAVINE-ADENINE DINUCLEOTIDE, Tryptophan 6-halogenase | | 著者 | Chitnumsub, P, Jaruwat, A, Phintha, A, Chaiyen, P. | | 登録日 | 2020-08-20 | | 公開日 | 2020-11-25 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Dissecting the low catalytic capability of flavin-dependent halogenases.
J.Biol.Chem., 296, 2020
|
|
