7PH3
 
 | | AMP-PNP bound nanodisc reconstituted MsbA with nanobodies, spin-labeled at position A60C | | 分子名称: | (1~{R},4~{R},11~{S},14~{S},19~{Z})-19-[2-[2,5-bis(oxidanylidene)pyrrolidin-1-yl]ethylimino]-7,8,17,18-tetraoxa-1,4,11,14-tetrazatricyclo[12.6.2.2^{4,11}]tetracosane-6,9,16-trione, 1,2-Distearoyl-sn-glycerophosphoethanolamine, ATP-dependent lipid A-core flippase, ... | | 著者 | Parey, K, Januliene, D, Galazzo, L, Meier, G, Vecchis, D, Striednig, B, Hilbi, H, Schaefer, L.V, Kuprov, I, Bordignon, E, Seeger, M.A, Moeller, A. | | 登録日 | 2021-08-16 | | 公開日 | 2022-08-24 | | 最終更新日 | 2022-10-26 | | 実験手法 | ELECTRON MICROSCOPY (2.8 Å) | | 主引用文献 | The ABC transporter MsbA adopts the wide inward-open conformation in E. coli cells.
Sci Adv, 8, 2022
|
|
7PXS
 
 | | Room temperature X-ray structure of LPMO at 1.91x10^3 Gy | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Auxiliary activity 9, COPPER (II) ION | | 著者 | Tandrup, T, Muderspach, S.J, Banerjee, S, Lo Leggio, L. | | 登録日 | 2021-10-08 | | 公開日 | 2022-08-24 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Changes in active-site geometry on X-ray photoreduction of a lytic polysaccharide monooxygenase active-site copper and saccharide binding.
Iucrj, 9, 2022
|
|
7PYQ
 
 | | Structure of an LPMO (expressed in E.coli) at 6.35x10^6 Gy | | 分子名称: | ACETATE ION, Auxiliary activity 9, COPPER (II) ION, ... | | 著者 | Tandrup, T, Muderspach, S.J, Banerjee, S, Ipsen, J.O, Rollan, C.H, Norholm, M.H.H, Johansen, K.S, Lo Leggio, L. | | 登録日 | 2021-10-10 | | 公開日 | 2022-08-24 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Changes in active-site geometry on X-ray photoreduction of a lytic polysaccharide monooxygenase active-site copper and saccharide binding.
Iucrj, 9, 2022
|
|
7PYE
 
 | |
7PYP
 
 | | Structure of an LPMO (expressed in E.coli) at 2.13x10^6 Gy | | 分子名称: | ACETATE ION, Auxiliary activity 9, COPPER (II) ION, ... | | 著者 | Tandrup, T, Muderspach, S.J, Banerjee, S, Ipsen, J.O, Rollan, C.H, Norholm, M.H.H, Johansen, K.S, Lo Leggio, L. | | 登録日 | 2021-10-10 | | 公開日 | 2022-08-24 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Changes in active-site geometry on X-ray photoreduction of a lytic polysaccharide monooxygenase active-site copper and saccharide binding.
Iucrj, 9, 2022
|
|
7PZ4
 
 | | Structure of an LPMO at 2.07x10^4 Gy | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ACRYLIC ACID, ... | | 著者 | Tandrup, T, Muderspach, S.J, Ipsen, J.O, Johansen, K.S, Lo Leggio, L. | | 登録日 | 2021-10-11 | | 公開日 | 2022-08-24 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Changes in active-site geometry on X-ray photoreduction of a lytic polysaccharide monooxygenase active-site copper and saccharide binding.
Iucrj, 9, 2022
|
|
7PZ6
 
 | | Structure of an LPMO at 2.22x10^5 Gy | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ACRYLIC ACID, ... | | 著者 | Tandrup, T, Muderspach, S.J, Ipsen, J.O, Johansen, K.S, Lo Leggio, L. | | 登録日 | 2021-10-11 | | 公開日 | 2022-08-24 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.45 Å) | | 主引用文献 | Changes in active-site geometry on X-ray photoreduction of a lytic polysaccharide monooxygenase active-site copper and saccharide binding.
Iucrj, 9, 2022
|
|
1OT4
 
 | | Solution structure of Cu(II)-CopC from Pseudomonas syringae | | 分子名称: | COPPER (II) ION, Copper resistance protein C | | 著者 | Arnesano, F, Banci, L, Bertini, I, Felli, I.C, Luchinat, C, Thompsett, A.R, Structural Proteomics in Europe (SPINE) | | 登録日 | 2003-03-21 | | 公開日 | 2003-07-08 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | A Strategy for the NMR Characterization of Type II Copper(II) Proteins:
the Case of the Copper Trafficking Protein CopC from Pseudomonas Syringae.
J.Am.Chem.Soc., 125, 2003
|
|
7PH7
 
 | | Nanodisc reconstituted MsbA in complex with nanobodies, spin-labeled at position T68C | | 分子名称: | (1~{R},4~{R},11~{S},14~{S},19~{Z})-19-[2-[2,5-bis(oxidanylidene)pyrrolidin-1-yl]ethylimino]-7,8,17,18-tetraoxa-1,4,11,14-tetrazatricyclo[12.6.2.2^{4,11}]tetracosane-6,9,16-trione, (2~{R},4~{R},5~{R},6~{R})-6-[(1~{R})-1,2-bis(oxidanyl)ethyl]-4-[(2~{R},3~{S},4~{S},5~{R},6~{R})-6-[(1~{S})-1,2-bis(oxidanyl)ethyl]-4-[(2~{R},3~{S},4~{S},5~{S},6~{R})-6-[(1~{S})-1,2-bis(oxidanyl)ethyl]-3,4,5-tris(oxidanyl)oxan-2-yl]oxy-3,5-bis(oxidanyl)oxan-2-yl]oxy-2-[[(2~{R},3~{S},4~{R},5~{R},6~{R})-4-[(3~{R})-3-nonanoyloxytetradecanoyl]oxy-5-[[(3~{R})-3-octanoyloxytetradecanoyl]amino]-6-[[(2~{R},3~{S},4~{S},5~{S},6~{R})-3-oxidanyl-5-[[(3~{R})-3-oxidanylnonanoyl]amino]-4-[(3~{R})-3-oxidanyltetradecanoyl]oxy-6-phosphonooxy-oxan-2-yl]methoxy]-3-phosphonooxy-oxan-2-yl]methoxy]-5-oxidanyl-oxane-2-carboxylic acid, ATP-binding transport protein multicopy suppressor of htrB, ... | | 著者 | Parey, K, Januliene, D, Galazzo, L, Meier, G, Vecchis, D, Striednig, B, Hilbi, H, Schaefer, L.V, Kuprov, I, Bordignon, E, Seeger, M.A, Moeller, A. | | 登録日 | 2021-08-16 | | 公開日 | 2022-08-24 | | 最終更新日 | 2022-10-26 | | 実験手法 | ELECTRON MICROSCOPY (4.1 Å) | | 主引用文献 | The ABC transporter MsbA adopts the wide inward-open conformation in E. coli cells.
Sci Adv, 8, 2022
|
|
7PYD
 
 | |
7PYY
 
 | | Structure of LPMO (expressed in E.coli) with cellotriose at 5.05x10^5 Gy | | 分子名称: | Auxiliary activity 9, CHLORIDE ION, COPPER (II) ION, ... | | 著者 | Tandrup, T, Muderspach, S.J, Banerjee, S, Ipsen, J.O, Rollan, C.H, Norholm, M.H.H, Johansen, K.S, Lo Leggio, L. | | 登録日 | 2021-10-11 | | 公開日 | 2022-08-24 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.2 Å) | | 主引用文献 | Changes in active-site geometry on X-ray photoreduction of a lytic polysaccharide monooxygenase active-site copper and saccharide binding.
Iucrj, 9, 2022
|
|
7PH2
 
 | | Nanodisc reconstituted MsbA in complex with nanobodies, spin-labeled at position A60C | | 分子名称: | (1~{R},4~{R},11~{S},14~{S},19~{Z})-19-[2-[2,5-bis(oxidanylidene)pyrrolidin-1-yl]ethylimino]-7,8,17,18-tetraoxa-1,4,11,14-tetrazatricyclo[12.6.2.2^{4,11}]tetracosane-6,9,16-trione, (2~{R},4~{R},5~{R},6~{R})-6-[(1~{R})-1,2-bis(oxidanyl)ethyl]-4-[(2~{R},3~{S},4~{S},5~{R},6~{R})-6-[(1~{S})-1,2-bis(oxidanyl)ethyl]-4-[(2~{R},3~{S},4~{S},5~{S},6~{R})-6-[(1~{S})-1,2-bis(oxidanyl)ethyl]-3,4,5-tris(oxidanyl)oxan-2-yl]oxy-3,5-bis(oxidanyl)oxan-2-yl]oxy-2-[[(2~{R},3~{S},4~{R},5~{R},6~{R})-4-[(3~{R})-3-nonanoyloxytetradecanoyl]oxy-5-[[(3~{R})-3-octanoyloxytetradecanoyl]amino]-6-[[(2~{R},3~{S},4~{S},5~{S},6~{R})-3-oxidanyl-5-[[(3~{R})-3-oxidanylnonanoyl]amino]-4-[(3~{R})-3-oxidanyltetradecanoyl]oxy-6-phosphonooxy-oxan-2-yl]methoxy]-3-phosphonooxy-oxan-2-yl]methoxy]-5-oxidanyl-oxane-2-carboxylic acid, ATP-dependent lipid A-core flippase, ... | | 著者 | Januliene, D, Parey, K, Galazzo, L, Meier, G, Vecchis, D, Striednig, B, Hilbi, H, Schaefer, L.V, Kuprov, I, Bordignon, E, Seeger, M.A, Moeller, A. | | 登録日 | 2021-08-16 | | 公開日 | 2022-08-24 | | 最終更新日 | 2022-10-26 | | 実験手法 | ELECTRON MICROSCOPY (3.7 Å) | | 主引用文献 | The ABC transporter MsbA adopts the wide inward-open conformation in E. coli cells.
Sci Adv, 8, 2022
|
|
7PYN
 
 | | Structure of an LPMO (expressed in E.coli) at 2.31x10^5 Gy | | 分子名称: | Auxiliary activity 9, COPPER (II) ION, SULFATE ION | | 著者 | Tandrup, T, Muderspach, S.J, Banerjee, S, Ipsen, J.O, Rollan, C.H, Norholm, M.H.H, Johansen, K.S, Lo Leggio, L. | | 登録日 | 2021-10-10 | | 公開日 | 2022-08-24 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | Changes in active-site geometry on X-ray photoreduction of a lytic polysaccharide monooxygenase active-site copper and saccharide binding.
Iucrj, 9, 2022
|
|
7PXR
 
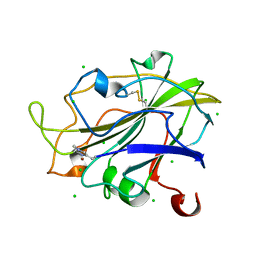 | | Room temperature structure of an LPMO. | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Auxiliary activity 9, CHLORIDE ION, ... | | 著者 | Tandrup, T, Meilleur, F, Ipsen, J, Johansen, K.S, Lo Leggio, L. | | 登録日 | 2021-10-08 | | 公開日 | 2022-08-24 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Changes in active-site geometry on X-ray photoreduction of a lytic polysaccharide monooxygenase active-site copper and saccharide binding.
Iucrj, 9, 2022
|
|
7PXJ
 
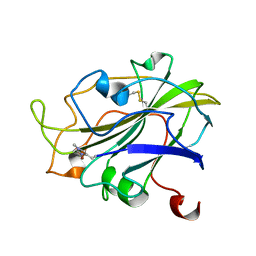 | | X-ray structure of LPMO at 5.99x10^4 Gy | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Auxiliary activity 9, COPPER (II) ION | | 著者 | Tandrup, T, Lo Leggio, L. | | 登録日 | 2021-10-08 | | 公開日 | 2022-08-24 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Changes in active-site geometry on X-ray photoreduction of a lytic polysaccharide monooxygenase active-site copper and saccharide binding.
Iucrj, 9, 2022
|
|
7PYI
 
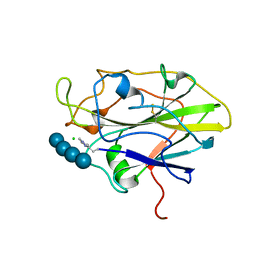 | |
7PYX
 
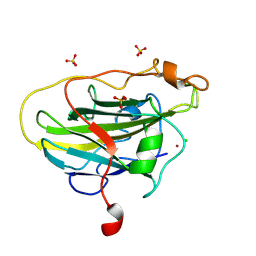 | | Structure of LPMO (expressed in E.coli) with cellotriose at 2.74x10^5 Gy | | 分子名称: | Auxiliary activity 9, CHLORIDE ION, COPPER (II) ION, ... | | 著者 | Tandrup, T, Muderspach, S.J, Banerjee, S, Ipsen, J.O, Rollan, C.H, Norholm, M.H.H, Johansen, K.S, Lo Leggio, L. | | 登録日 | 2021-10-11 | | 公開日 | 2022-08-24 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Changes in active-site geometry on X-ray photoreduction of a lytic polysaccharide monooxygenase active-site copper and saccharide binding.
Iucrj, 9, 2022
|
|
1ORW
 
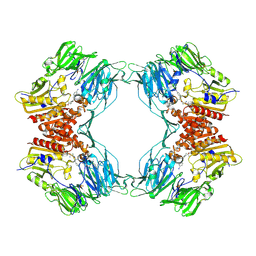 | | Crystal Structure of Porcine Dipeptidyl Peptidase IV (CD26) in Complex with a Peptidomimetic Inhibitor | | 分子名称: | (2S)-PYRROLIDIN-2-YLMETHYLAMINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Engel, M, Hoffmann, T, Wagner, L, Wermann, M, Heiser, U, Kiefersauer, R, Huber, R, Bode, W, Demuth, H.U, Brandstetter, H. | | 登録日 | 2003-03-16 | | 公開日 | 2003-05-06 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (2.84 Å) | | 主引用文献 | The Crystal Structure of Dipeptidyl Peptidase IV (CD26) Reveals its Functional Regulation and Enzymatic Mechanism
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
7PZ3
 
 | | Structure of an LPMO at 5.37x10^3 Gy | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ACRYLIC ACID, ... | | 著者 | Tandrup, T, Muderspach, S.J, Ipsen, J.O, Johansen, K.S, Lo Leggio, L. | | 登録日 | 2021-10-11 | | 公開日 | 2022-08-24 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Changes in active-site geometry on X-ray photoreduction of a lytic polysaccharide monooxygenase active-site copper and saccharide binding.
Iucrj, 9, 2022
|
|
7PFM
 
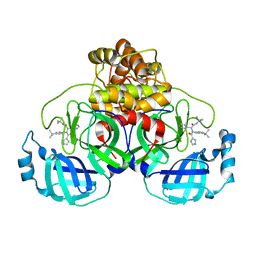 | | A SARS-CoV2 major protease non-covalent ligand structure determined to 2.0 A resolution | | 分子名称: | N-[(1R)-2-(tert-butylamino)-2-oxidanylidene-1-pyridin-3-yl-ethyl]-N-(4-tert-butylphenyl)-1H-imidazole-5-carboxamide, Replicase polyprotein 1ab | | 著者 | Moche, M, Moodie, L, Strandback, E, Nyman, T, Sandstrom, A, Akaberi, D, Lennerstrand, J. | | 登録日 | 2021-08-11 | | 公開日 | 2022-08-24 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | The SARS-CoV2 major protease (Mpro) in complex with a non-covalent inhibitory ligand at 2 A resolution
To Be Published
|
|
7PH4
 
 | | AMP-PNP bound nanodisc reconstituted MsbA with nanobodies, spin-labeled at position T68C | | 分子名称: | (1~{R},4~{R},11~{S},14~{S},19~{Z})-19-[2-[2,5-bis(oxidanylidene)pyrrolidin-1-yl]ethylimino]-7,8,17,18-tetraoxa-1,4,11,14-tetrazatricyclo[12.6.2.2^{4,11}]tetracosane-6,9,16-trione, ATP-dependent lipid A-core flippase, DODECYL-BETA-D-MALTOSIDE, ... | | 著者 | Parey, K, Januliene, D, Galazzo, L, Meier, G, Vecchis, D, Striednig, B, Hilbi, H, Schaefer, L.V, Kuprov, I, Bordignon, E, Seeger, M.A, Moeller, A. | | 登録日 | 2021-08-16 | | 公開日 | 2022-08-24 | | 最終更新日 | 2022-10-26 | | 実験手法 | ELECTRON MICROSCOPY (2.8 Å) | | 主引用文献 | The ABC transporter MsbA adopts the wide inward-open conformation in E. coli cells.
Sci Adv, 8, 2022
|
|
7PXK
 
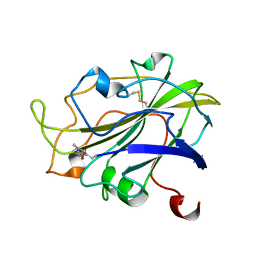 | | X-ray structure of LPMO at 1.39x10^5 Gy | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Auxiliary activity 9, COPPER (II) ION | | 著者 | Tandrup, T, Lo Leggio, L. | | 登録日 | 2021-10-08 | | 公開日 | 2022-08-24 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | Changes in active-site geometry on X-ray photoreduction of a lytic polysaccharide monooxygenase active-site copper and saccharide binding.
Iucrj, 9, 2022
|
|
4E8F
 
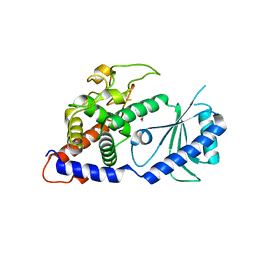 | | Structural Basis for the Activity of a Cytoplasmic RNA Terminal U-transferase | | 分子名称: | ACETATE ION, GLYCEROL, Poly(A) RNA polymerase protein cid1 | | 著者 | Yates, L.A, Fleurdepine, S, Rissland, O.S, DeColibus, L, Harlos, K, Norbury, C.J, Gilbert, R.J.C. | | 登録日 | 2012-03-20 | | 公開日 | 2012-07-04 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Structural basis for the activity of a cytoplasmic RNA terminal uridylyl transferase.
Nat.Struct.Mol.Biol., 19, 2012
|
|
7PXU
 
 | | LsAA9_A chemically reduced with ascorbic acid (low X-ray dose) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Auxiliary activity 9, CHLORIDE ION, ... | | 著者 | Tandrup, T, Muderspach, S.J, Banerjee, S, Ipsen, J, Johansen, K.S, Lo Leggio, L. | | 登録日 | 2021-10-08 | | 公開日 | 2022-08-24 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Changes in active-site geometry on X-ray photoreduction of a lytic polysaccharide monooxygenase active-site copper and saccharide binding.
Iucrj, 9, 2022
|
|
7PYM
 
 | | Structure of an LPMO (expressed in E.coli) at 5.61x10^4 Gy | | 分子名称: | Auxiliary activity 9, COPPER (II) ION, SULFATE ION | | 著者 | Tandrup, T, Muderspach, S.J, Banerjee, S, Ipsen, J.O, Rollan, C.H, Norholm, M.H.H, Johansen, K.S, Lo Leggio, L. | | 登録日 | 2021-10-10 | | 公開日 | 2022-08-24 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Changes in active-site geometry on X-ray photoreduction of a lytic polysaccharide monooxygenase active-site copper and saccharide binding.
Iucrj, 9, 2022
|
|
