3SHY
 
 | | Crystal structure of the PDE5A1 catalytic domain in complex with novel inhibitors | | 分子名称: | 6-ethyl-5-fluoro-2-{5-[(4-methylpiperazin-1-yl)sulfonyl]-2-propoxyphenyl}pyrimidin-4(3H)-one, MAGNESIUM ION, ZINC ION, ... | | 著者 | Chen, T.T, Chen, T, Xu, Y.C. | | 登録日 | 2011-06-17 | | 公開日 | 2011-08-24 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.647 Å) | | 主引用文献 | Utilization of Halogen Bond in Lead Optimization: A Case Study of Rational Design of Potent Phosphodiesterase Type 5 (PDE5) Inhibitors.
J.Med.Chem., 54, 2011
|
|
7BXF
 
 | | MvcA-Lpg2149 complex | | 分子名称: | Lpg2149, MvcA, PRASEODYMIUM ION | | 著者 | Gao, P. | | 登録日 | 2020-04-19 | | 公開日 | 2020-05-20 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Insights into catalysis and regulation of non-canonical ubiquitination and deubiquitination by bacterial deamidase effectors.
Nat Commun, 11, 2020
|
|
7KN3
 
 | | Crystal structure of SARS-CoV-2 spike protein receptor-binding domain complexed with a pre-pandemic antibody S-B8 Fab | | 分子名称: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, S-B8 Fab heavy chain, ... | | 著者 | Liu, H, Zhu, X, Wilson, I.A. | | 登録日 | 2020-11-04 | | 公開日 | 2021-09-22 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.251 Å) | | 主引用文献 | Neutralizing Antibodies to SARS-CoV-2 Selected from a Human Antibody Library Constructed Decades Ago.
Adv Sci, 9, 2022
|
|
7KN4
 
 | |
5E06
 
 | |
3SIE
 
 | |
5OFL
 
 | | Crystal structure of CbXyn10C variant E140Q/E248Q complexed with cellohexaose | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, Glycoside hydrolase family 48, SULFATE ION, ... | | 著者 | Hakulinen, N, Penttinen, L, Rouvinen, J. | | 登録日 | 2017-07-11 | | 公開日 | 2017-10-04 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.871 Å) | | 主引用文献 | Insights into the roles of non-catalytic residues in the active site of a GH10 xylanase with activity on cellulose.
J. Biol. Chem., 292, 2017
|
|
5OFK
 
 | | Crystal structure of CbXyn10C variant E140Q/E248Q complexed with xyloheptaose | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, Glycoside hydrolase family 48, PIPERAZINE-N,N'-BIS(2-ETHANESULFONIC ACID), ... | | 著者 | Hakulinen, N, Penttinen, L, Rouvinen, J, Tu, T. | | 登録日 | 2017-07-11 | | 公開日 | 2017-10-04 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.16 Å) | | 主引用文献 | Insights into the roles of non-catalytic residues in the active site of a GH10 xylanase with activity on cellulose.
J. Biol. Chem., 292, 2017
|
|
5OFJ
 
 | | Crystal structure of N-terminal domain of bifunctional CbXyn10C | | 分子名称: | 1,2-ETHANEDIOL, CITRATE ANION, Glycoside hydrolase family 48 | | 著者 | Hakulinen, N, Penttinen, L, Rouvinen, J. | | 登録日 | 2017-07-11 | | 公開日 | 2017-10-04 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.34 Å) | | 主引用文献 | Insights into the roles of non-catalytic residues in the active site of a GH10 xylanase with activity on cellulose.
J. Biol. Chem., 292, 2017
|
|
7SEL
 
 | | E. coli MsbA in complex with LPS and inhibitor G7090 (compound 3) | | 分子名称: | (2E)-3-{7-[(1S)-1-(2,6-dichloro-3-fluorophenyl)ethoxy]-1-methylnaphthalen-2-yl}prop-2-enoic acid, (2~{R},4~{R},5~{R},6~{R})-6-[(1~{R})-1,2-bis(oxidanyl)ethyl]-2-[(2~{R},4~{R},5~{R},6~{R})-6-[(1~{R})-1,2-bis(oxidanyl)ethyl]-2-carboxy-2-[[(2~{R},3~{S},4~{R},5~{R},6~{R})-5-[[(3~{R})-3-dodecanoyloxytetradecanoyl]amino]-6-[[(2~{R},3~{S},4~{R},5~{R},6~{R})-3-oxidanyl-5-[[(3~{R})-3-oxidanyltetradecanoyl]amino]-4-[(3~{R})-3-oxidanyltetradecanoyl]oxy-6-phosphonooxy-oxan-2-yl]methoxy]-3-phosphonooxy-4-[(3~{R})-3-tetradecanoyloxytetradecanoyl]oxy-oxan-2-yl]methoxy]-5-oxidanyl-oxan-4-yl]oxy-4,5-bis(oxidanyl)oxane-2-carboxylic acid, ATP-dependent lipid A-core flippase | | 著者 | Payandeh, J, Koth, C.M, Verma, V.A. | | 登録日 | 2021-09-30 | | 公開日 | 2022-03-09 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (2.978 Å) | | 主引用文献 | Discovery of Inhibitors of the Lipopolysaccharide Transporter MsbA: From a Screening Hit to Potent Wild-Type Gram-Negative Activity.
J.Med.Chem., 65, 2022
|
|
8DKC
 
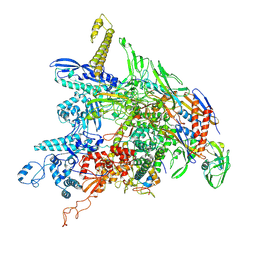 | | P. gingivalis RNA Polymerase | | 分子名称: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | 著者 | Liu, B, Bu, F. | | 登録日 | 2022-07-05 | | 公開日 | 2023-06-21 | | 最終更新日 | 2024-05-01 | | 実験手法 | ELECTRON MICROSCOPY (3.5 Å) | | 主引用文献 | Cryo-EM Structure of Porphyromonas gingivalis RNA Polymerase.
J.Mol.Biol., 436, 2024
|
|
4TTH
 
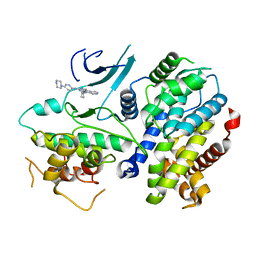 | | Crystal structure of a CDK6/Vcyclin complex with inhibitor bound | | 分子名称: | 9-cyclopentyl-N-(5-piperazin-1-ylpyridin-2-yl)pyrido[4,5]pyrrolo[1,2-d]pyrimidin-2-amine, Cyclin homolog, Cyclin-dependent kinase 6 | | 著者 | Piper, D.E, Walker, N, Wang, Z. | | 登録日 | 2014-06-20 | | 公開日 | 2014-08-06 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Discovery of AMG 925, a FLT3 and CDK4 dual kinase inhibitor with preferential affinity for the activated state of FLT3.
J.Med.Chem., 57, 2014
|
|
3AJM
 
 | | Crystal structure of programmed cell death 10 in complex with inositol 1,3,4,5-tetrakisphosphate | | 分子名称: | INOSITOL-(1,3,4,5)-TETRAKISPHOSPHATE, Programmed cell death protein 10 | | 著者 | Ding, J, Wang, D.C. | | 登録日 | 2010-06-09 | | 公開日 | 2010-06-30 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Crystal structure of human programmed cell death 10 complexed with inositol-(1,3,4,5)-tetrakisphosphate: a novel adaptor protein involved in human cerebral cavernous malformation.
Biochem.Biophys.Res.Commun., 399, 2010
|
|
6A50
 
 | | structure of benzoylformate decarboxylases in complex with cofactor TPP | | 分子名称: | MAGNESIUM ION, THIAMINE DIPHOSPHATE, benzoylformate decarboxylases | | 著者 | Guo, Y, Wang, S, Nie, Y, Li, S. | | 登録日 | 2018-06-21 | | 公開日 | 2019-02-20 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | A Synthetic Pathway for Acetyl-Coenzyme A Biosynthesis
Nat Commun, 2019
|
|
6M6I
 
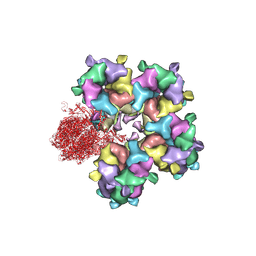 | | Structure of HSV2 B-capsid portal vertex | | 分子名称: | Coiled coils chain 1, Coiled coils chain 2, Major capsid protein, ... | | 著者 | Wang, X.X, Wang, N. | | 登録日 | 2020-03-14 | | 公開日 | 2021-03-10 | | 実験手法 | ELECTRON MICROSCOPY (4.05 Å) | | 主引用文献 | Structures of the portal vertex reveal essential protein-protein interactions for Herpesvirus assembly and maturation.
Protein Cell, 11, 2020
|
|
6M6H
 
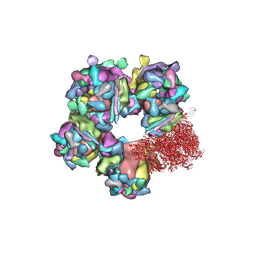 | | Structure of HSV2 C-capsid portal vertex | | 分子名称: | Capsid vertex component 1, Capsid vertex component 2, Large tegument protein deneddylase, ... | | 著者 | Wang, X.X, Wang, N. | | 登録日 | 2020-03-14 | | 公開日 | 2021-03-24 | | 実験手法 | ELECTRON MICROSCOPY (4.5 Å) | | 主引用文献 | Structures of the portal vertex reveal essential protein-protein interactions for Herpesvirus assembly and maturation.
Protein Cell, 11, 2020
|
|
6M6G
 
 | | Structure of HSV2 viron capsid portal vertex | | 分子名称: | Capsid vertex component 1, Capsid vertex component 2, Coiled coils, ... | | 著者 | Wang, X.X, Wang, N. | | 登録日 | 2020-03-14 | | 公開日 | 2021-03-24 | | 実験手法 | ELECTRON MICROSCOPY (5.39 Å) | | 主引用文献 | Structures of the portal vertex reveal essential protein-protein interactions for Herpesvirus assembly and maturation.
Protein Cell, 11, 2020
|
|
2KLJ
 
 | | Solution Structure of gammaD-Crystallin with RDC and SAXS | | 分子名称: | Gamma-crystallin D | | 著者 | Wang, J, Zuo, X, Yu, P, Byeon, I, Jung, J, Gronenborn, A.M, Wang, Y. | | 登録日 | 2009-07-06 | | 公開日 | 2009-10-06 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR, SOLUTION SCATTERING | | 主引用文献 | Determination of multicomponent protein structures in solution using global orientation and shape restraints.
J.Am.Chem.Soc., 131, 2009
|
|
2KLM
 
 | | Solution Structure of L11 with SAXS and RDC | | 分子名称: | 50S ribosomal protein L11 | | 著者 | Wang, J, Zuo, X, Yu, P, Schwieters, C.D, Wang, Y. | | 登録日 | 2009-07-06 | | 公開日 | 2009-10-06 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR, SOLUTION SCATTERING | | 主引用文献 | Determination of multicomponent protein structures in solution using global orientation and shape restraints.
J.Am.Chem.Soc., 131, 2009
|
|
4TVZ
 
 | | Crystal Structure of SCARB2 in Neural Condition (pH7.5) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Scavenger receptor class B member 2, ... | | 著者 | Dang, M.H, Wang, X.X, Rao, Z.H. | | 登録日 | 2014-06-29 | | 公開日 | 2015-07-08 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (3.006 Å) | | 主引用文献 | Molecular mechanism of SCARB2-mediated attachment and uncoating of EV71
Protein Cell, 5, 2014
|
|
5ZZ8
 
 | | Structure of the Herpes simplex virus type 2 C-capsid with capsid-vertex-specific component | | 分子名称: | Major capsid protein, UL17, UL25, ... | | 著者 | Wang, J.L, Yuan, S, Zhu, D.J, Tang, H, Wang, N, Chen, W.Y, Gao, Q, Li, Y.H, Wang, J.Z, Liu, H.R, Zhang, X.Z, Rao, Z.H, Wang, X.X. | | 登録日 | 2018-05-31 | | 公開日 | 2018-10-10 | | 最終更新日 | 2019-11-06 | | 実験手法 | ELECTRON MICROSCOPY (3.75 Å) | | 主引用文献 | Structure of the herpes simplex virus type 2 C-capsid with capsid-vertex-specific component.
Nat Commun, 9, 2018
|
|
7XYT
 
 | |
6IVE
 
 | |
4LN0
 
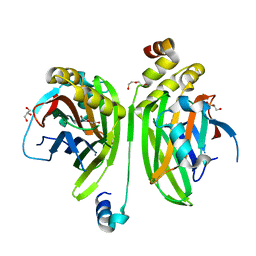 | | Crystal structure of the VGLL4-TEAD4 complex | | 分子名称: | DI(HYDROXYETHYL)ETHER, GLYCEROL, Transcription cofactor vestigial-like protein 4, ... | | 著者 | Wang, H, Shi, Z, Zhou, Z. | | 登録日 | 2013-07-11 | | 公開日 | 2014-02-26 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.896 Å) | | 主引用文献 | A Peptide Mimicking VGLL4 Function Acts as a YAP Antagonist Therapy against Gastric Cancer.
Cancer Cell, 25, 2014
|
|
3A9U
 
 | |
