3K4T
 
 | |
4WOI
 
 | | 4,5-linked aminoglycoside antibiotics regulate the bacterial ribosome by targeting dynamic conformational processes within intersubunit bridge B2 | | 分子名称: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | 著者 | Pulk, A, Cate, J.H.D, Blanchard, S, Wasserman, M, Altman, R, Zhou, Z, Zinder, J, Green, K, Garneau-Tsodikova, S. | | 登録日 | 2014-10-15 | | 公開日 | 2015-08-05 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Chemically related 4,5-linked aminoglycoside antibiotics drive subunit rotation in opposite directions.
Nat Commun, 6, 2015
|
|
7RMT
 
 | | Room temperature X-ray structure of SARS-CoV-2 main protease (Mpro) in complex with HL-3-70 | | 分子名称: | 2-chloro-4-[4-(2,6-dioxo-1,2,5,6-tetrahydropyrimidine-4-carbonyl)piperazin-1-yl]benzaldehyde, 3C-like proteinase | | 著者 | Kovalevsky, A, Kneller, D.W, Coates, L. | | 登録日 | 2021-07-28 | | 公開日 | 2021-11-10 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structural, Electronic, and Electrostatic Determinants for Inhibitor Binding to Subsites S1 and S2 in SARS-CoV-2 Main Protease.
J.Med.Chem., 64, 2021
|
|
7RNH
 
 | | Room temperature X-ray structure of SARS-CoV-2 main protease (Mpro) in complex with HL-3-45 | | 分子名称: | 3C-like proteinase, 6-[4-(4-chlorophenyl)piperazine-1-carbonyl]pyrimidine-2,4(1H,3H)-dione | | 著者 | Kovalevsky, A, Kneller, D.W, Coates, L. | | 登録日 | 2021-07-29 | | 公開日 | 2021-11-10 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structural, Electronic, and Electrostatic Determinants for Inhibitor Binding to Subsites S1 and S2 in SARS-CoV-2 Main Protease.
J.Med.Chem., 64, 2021
|
|
7RLS
 
 | | Room temperature X-ray structure of SARS-CoV-2 main protease (Mpro) in complex with HL-3-68 | | 分子名称: | 3C-like proteinase, 6-[4-(3,4,5-trichlorophenyl)piperazine-1-carbonyl]pyrimidine-2,4(1H,3H)-dione | | 著者 | Kovalevsky, A, Kneller, D.W, Coates, L. | | 登録日 | 2021-07-26 | | 公開日 | 2021-11-10 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structural, Electronic, and Electrostatic Determinants for Inhibitor Binding to Subsites S1 and S2 in SARS-CoV-2 Main Protease.
J.Med.Chem., 64, 2021
|
|
7RNK
 
 | | Room temperature X-ray structure of SARS-CoV-2 main protease (Mpro) in complex with HL-3-71 | | 分子名称: | 3C-like proteinase, 6-{4-[3-chloro-4-(hydroxymethyl)phenyl]piperazine-1-carbonyl}pyrimidine-2,4(3H,5H)-dione | | 著者 | Kovalevsky, A, Kneller, D.W, Coates, L. | | 登録日 | 2021-07-29 | | 公開日 | 2021-11-10 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Structural, Electronic, and Electrostatic Determinants for Inhibitor Binding to Subsites S1 and S2 in SARS-CoV-2 Main Protease.
J.Med.Chem., 64, 2021
|
|
7RM2
 
 | | Room temperature X-ray structure of SARS-CoV-2 main protease (Mpro) in complex with Mcule-CSR-494190-S1 | | 分子名称: | 3C-like proteinase, 6-[4-(3,5-dichloro-4-methylphenyl)piperazine-1-carbonyl]pyrimidine-2,4(1H,3H)-dione | | 著者 | Kovalevsky, A, Kneller, D.W, Coates, L. | | 登録日 | 2021-07-26 | | 公開日 | 2021-11-10 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structural, Electronic, and Electrostatic Determinants for Inhibitor Binding to Subsites S1 and S2 in SARS-CoV-2 Main Protease.
J.Med.Chem., 64, 2021
|
|
7RME
 
 | | Room temperature X-ray structure of SARS-CoV-2 main protease (Mpro) in complex with HL-3-52 | | 分子名称: | 3C-like proteinase, 6-{4-[4-chloro-3-(trifluoromethyl)phenyl]piperazine-1-carbonyl}pyrimidine-2,4(1H,3H)-dione | | 著者 | Kovalevsky, A, Kneller, D.W, Coates, L. | | 登録日 | 2021-07-27 | | 公開日 | 2021-11-10 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structural, Electronic, and Electrostatic Determinants for Inhibitor Binding to Subsites S1 and S2 in SARS-CoV-2 Main Protease.
J.Med.Chem., 64, 2021
|
|
7RMB
 
 | | Room temperature X-ray structure of SARS-CoV-2 main protease (Mpro) in complex with HL-3-78 | | 分子名称: | 3C-like proteinase, 6-[4-(4-bromo-3-chlorophenyl)piperazine-1-carbonyl]pyrimidine-2,4(1H,3H)-dione | | 著者 | Kovalevsky, A, Kneller, D.W, Coates, L. | | 登録日 | 2021-07-27 | | 公開日 | 2021-11-10 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structural, Electronic, and Electrostatic Determinants for Inhibitor Binding to Subsites S1 and S2 in SARS-CoV-2 Main Protease.
J.Med.Chem., 64, 2021
|
|
7RN4
 
 | | Room temperature X-ray structure of SARS-CoV-2 main protease (Mpro) in complex with HL-3-69 | | 分子名称: | 3C-like proteinase, 6-[4-(3,4-dichlorophenyl)piperidine-1-carbonyl]pyrimidine-2,4(1H,3H)-dione | | 著者 | Kovalevsky, A, Kneller, D.W, Coates, L. | | 登録日 | 2021-07-29 | | 公開日 | 2021-11-10 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Structural, Electronic, and Electrostatic Determinants for Inhibitor Binding to Subsites S1 and S2 in SARS-CoV-2 Main Protease.
J.Med.Chem., 64, 2021
|
|
3LEA
 
 | |
5FCS
 
 | | Diabody | | 分子名称: | Diabody, SULFATE ION | | 著者 | Mosyak, L, Root, A. | | 登録日 | 2015-12-15 | | 公開日 | 2016-12-14 | | 最終更新日 | 2019-11-13 | | 実験手法 | X-RAY DIFFRACTION (2.01 Å) | | 主引用文献 | Development of PF-06671008, a Highly Potent Anti-P-cadherin/Anti-CD3 Bispecific DART Molecule with Extended Half-Life for the Treatment of Cancer.
Antibodies, 5, 2016
|
|
5HG9
 
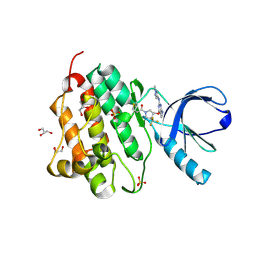 | | EGFR (L858R, T790M, V948R) in complex with 1-[(3R,4R)-3-[({2-[(1-methyl-1H-pyrazol-4-yl)amino]-7H-pyrrolo[2,3-d]pyrimidin-4-yl}oxy)methyl]-4-(trifluoromethyl)pyrrolidin-1-yl]prop-2-en-1-one | | 分子名称: | 1-[(3R,4R)-3-[({2-[(1-methyl-1H-pyrazol-4-yl)amino]-7H-pyrrolo[2,3-d]pyrimidin-4-yl}oxy)methyl]-4-(trifluoromethyl)pyrrolidin-1-yl]propan-1-one, Epidermal growth factor receptor, GLYCEROL, ... | | 著者 | Gajiwala, K.S. | | 登録日 | 2016-01-08 | | 公開日 | 2016-02-03 | | 最終更新日 | 2016-03-23 | | 実験手法 | X-RAY DIFFRACTION (2.15 Å) | | 主引用文献 | Discovery of 1-{(3R,4R)-3-[({5-Chloro-2-[(1-methyl-1H-pyrazol-4-yl)amino]-7H-pyrrolo[2,3-d]pyrimidin-4-yl}oxy)methyl]-4-methoxypyrrolidin-1-yl}prop-2-en-1-one (PF-06459988), a Potent, WT Sparing, Irreversible Inhibitor of T790M-Containing EGFR Mutants.
J.Med.Chem., 59, 2016
|
|
5HG8
 
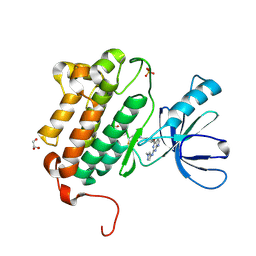 | | EGFR (L858R, T790M, V948R) in complex with N-[3-({2-[(1-methyl-1H-pyrazol-4-yl)amino]-7H-pyrrolo[2,3-d]pyrimidin-4-yl}oxy)phenyl]prop-2-enamide | | 分子名称: | Epidermal growth factor receptor, GLYCEROL, N-[3-({2-[(1-methyl-1H-pyrazol-4-yl)amino]-7H-pyrrolo[2,3-d]pyrimidin-4-yl}oxy)phenyl]propanamide, ... | | 著者 | Gajiwala, K.S. | | 登録日 | 2016-01-08 | | 公開日 | 2016-02-03 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (1.42 Å) | | 主引用文献 | Discovery of 1-{(3R,4R)-3-[({5-Chloro-2-[(1-methyl-1H-pyrazol-4-yl)amino]-7H-pyrrolo[2,3-d]pyrimidin-4-yl}oxy)methyl]-4-methoxypyrrolidin-1-yl}prop-2-en-1-one (PF-06459988), a Potent, WT Sparing, Irreversible Inhibitor of T790M-Containing EGFR Mutants.
J.Med.Chem., 59, 2016
|
|
3HMV
 
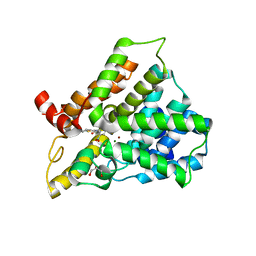 | | Catalytic domain of human phosphodiesterase 4B2B in complex with a tetrahydrobenzothiophene inhibitor | | 分子名称: | (6S)-6-methyl-2-{[(2-nitrophenyl)carbonyl]amino}-4,5,6,7-tetrahydro-1-benzothiophene-3-carboxamide, GLYCEROL, MAGNESIUM ION, ... | | 著者 | Somers, D.O, Neu, M. | | 登録日 | 2009-05-29 | | 公開日 | 2010-06-09 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.23 Å) | | 主引用文献 | Identification of PDE4B Over 4D subtype-selective inhibitors revealing an unprecedented binding mode
Bioorg.Med.Chem., 17, 2009
|
|
2GWX
 
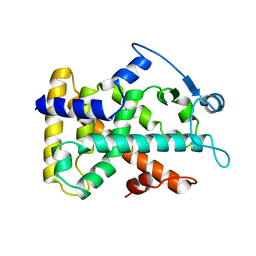 | | MOLECULAR RECOGNITION OF FATTY ACIDS BY PEROXISOME PROLIFERATOR-ACTIVATED RECEPTORS | | 分子名称: | PROTEIN (PPAR-DELTA) | | 著者 | Xu, H.E, Lambert, M.H, Montana, V.G, Park, D.J, Blanchard, S, Brown, P, Sternbach, D, Lehmann, J, Bruce, G.W, Willson, T.M, Kliewer, S.A, Milburn, M.V. | | 登録日 | 1999-03-11 | | 公開日 | 2000-03-11 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Molecular recognition of fatty acids by peroxisome proliferator-activated receptors.
Mol.Cell, 3, 1999
|
|
3BRJ
 
 | | Crystal structure of mannitol operon repressor (MtlR) from Vibrio parahaemolyticus RIMD 2210633 | | 分子名称: | 1,2-ETHANEDIOL, GLYCEROL, Mannitol operon repressor | | 著者 | Tan, K, Zhou, M, Moy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2007-12-21 | | 公開日 | 2008-01-15 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (2.75 Å) | | 主引用文献 | The mannitol operon repressor MtlR belongs to a new class of transcription regulators in bacteria.
J.Biol.Chem., 284, 2009
|
|
5OUG
 
 | | Humanized alpha-AChBP (acetylcholine binding protein) in complex with lobeline and allosteric binder fragment 4. | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 4,5-dibromo-N-(3-hydroxypropyl)-1H-pyrrole-2-carboxamide, Alpha-Lobeline, ... | | 著者 | Delbart, F, Gruss, F, Ulens, C. | | 登録日 | 2017-08-23 | | 公開日 | 2017-12-20 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (2.57 Å) | | 主引用文献 | An allosteric binding site of the alpha 7 nicotinic acetylcholine receptor revealed in a humanized acetylcholine-binding protein.
J. Biol. Chem., 293, 2018
|
|
3L0V
 
 | |
5HG7
 
 | | EGFR (L858R, T790M, V948R) in complex with 1-{(3R,4R)-3-[5-Chloro-2-(1-methyl-1H-pyrazol-4-ylamino)-7H-pyrrolo[2,3-d]pyrimidin-4-yloxymethyl]-4-methoxy-pyrrolidin-1-yl}propenone (PF-06459988) | | 分子名称: | 1-{(3R,4R)-3-[({5-chloro-2-[(1-methyl-1H-pyrazol-4-yl)amino]-7H-pyrrolo[2,3-d]pyrimidin-4-yl}oxy)methyl]-4-methoxypyrrolidin-1-yl}propan-1-one, Epidermal growth factor receptor, SULFATE ION | | 著者 | Gajiwala, K.S. | | 登録日 | 2016-01-08 | | 公開日 | 2016-01-27 | | 最終更新日 | 2016-03-23 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Discovery of 1-{(3R,4R)-3-[({5-Chloro-2-[(1-methyl-1H-pyrazol-4-yl)amino]-7H-pyrrolo[2,3-d]pyrimidin-4-yl}oxy)methyl]-4-methoxypyrrolidin-1-yl}prop-2-en-1-one (PF-06459988), a Potent, WT Sparing, Irreversible Inhibitor of T790M-Containing EGFR Mutants.
J.Med.Chem., 59, 2016
|
|
5OUI
 
 | | Humanized alpha-AChBP (acetylcholine binding protein) in complex with allosteric binder fragment CU2017 | | 分子名称: | (3~{R})-~{N}-(5-bromanylpyridin-2-yl)piperidine-3-carboxamide, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Delbart, F, Gruss, F, Ulens, C. | | 登録日 | 2017-08-23 | | 公開日 | 2017-11-29 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (3.1 Å) | | 主引用文献 | An allosteric binding site of the alpha 7 nicotinic acetylcholine receptor revealed in a humanized acetylcholine-binding protein.
J. Biol. Chem., 293, 2018
|
|
5FHA
 
 | |
5FHB
 
 | |
4P0I
 
 | | Structure of the PBP NocT | | 分子名称: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, Nopaline-binding periplasmic protein | | 著者 | Vigouroux, A, Morera, S. | | 登録日 | 2014-02-21 | | 公開日 | 2014-10-22 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.89 Å) | | 主引用文献 | Agrobacterium uses a unique ligand-binding mode for trapping opines and acquiring a competitive advantage in the niche construction on plant host.
Plos Pathog., 10, 2014
|
|
3LE9
 
 | |
