5CMZ
 
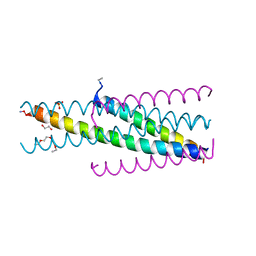 | | Artificial HIV fusion inhibitor AP3 fused to the C-terminus of gp41 NHR | | 分子名称: | 1,2-ETHANEDIOL, 1-ETHOXY-2-(2-ETHOXYETHOXY)ETHANE, Artificial HIV entry inhibitor AP3, ... | | 著者 | Zhu, Y, Ye, S, Zhang, R. | | 登録日 | 2015-07-17 | | 公開日 | 2015-09-16 | | 実験手法 | X-RAY DIFFRACTION (2.574 Å) | | 主引用文献 | Improved Pharmacological and Structural Properties of HIV Fusion Inhibitor AP3 over Enfuvirtide: Highlighting Advantages of Artificial Peptide Strategy.
Sci Rep, 5, 2015
|
|
8CZI
 
 | |
8D9C
 
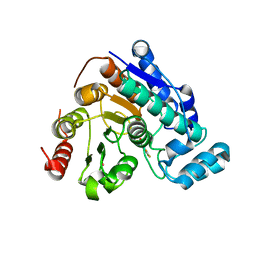 | | Crystal Structure of Danio rerio histone deacetylase 6 catalytic domain 2 complexed with fluorinated inhibitor 10 | | 分子名称: | 2,3,4,5,6-pentafluoro-N-hydroxybenzamide, Hdac6 protein, POTASSIUM ION, ... | | 著者 | Watson, P.R, Christianson, D.W. | | 登録日 | 2022-06-09 | | 公開日 | 2022-09-28 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.82 Å) | | 主引用文献 | Aromatic Ring Fluorination Patterns Modulate Inhibitory Potency of Fluorophenylhydroxamates Complexed with Histone Deacetylase 6.
Biochemistry, 61, 2022
|
|
8D99
 
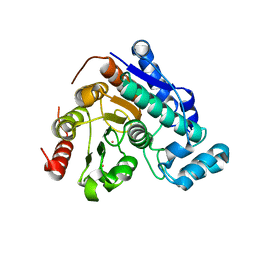 | | Crystal Structure of Danio rerio histone deacetylase 6 catalytic domain 2 complexed with fluorinated inhibitor 7 | | 分子名称: | 2,3,6-trifluoro-N-hydroxybenzamide, Hdac6 protein, POTASSIUM ION, ... | | 著者 | Watson, P.R, Christianson, D.W. | | 登録日 | 2022-06-09 | | 公開日 | 2022-09-28 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.79 Å) | | 主引用文献 | Aromatic Ring Fluorination Patterns Modulate Inhibitory Potency of Fluorophenylhydroxamates Complexed with Histone Deacetylase 6.
Biochemistry, 61, 2022
|
|
8D9B
 
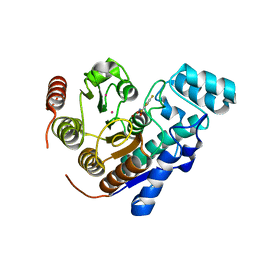 | | Crystal Structure of Danio rerio histone deacetylase 6 catalytic domain 2 complexed with fluorinated inhibitor 9 | | 分子名称: | 2,3,5,6-tetrafluoro-N-hydroxybenzamide, Hdac6 protein, POTASSIUM ION, ... | | 著者 | Watson, P.R, Christianson, D.W. | | 登録日 | 2022-06-09 | | 公開日 | 2022-09-28 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.63 Å) | | 主引用文献 | Aromatic Ring Fluorination Patterns Modulate Inhibitory Potency of Fluorophenylhydroxamates Complexed with Histone Deacetylase 6.
Biochemistry, 61, 2022
|
|
8D9A
 
 | | Crystal Structure of Danio rerio histone deacetylase 6 catalytic domain 2 complexed with fluorinated inhibitor 8 | | 分子名称: | 2,3,5-trifluoro-N-hydroxybenzamide, Hdac6 protein, POTASSIUM ION, ... | | 著者 | Watson, P.R, Christianson, D.W. | | 登録日 | 2022-06-09 | | 公開日 | 2022-09-28 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Aromatic Ring Fluorination Patterns Modulate Inhibitory Potency of Fluorophenylhydroxamates Complexed with Histone Deacetylase 6.
Biochemistry, 61, 2022
|
|
8D98
 
 | |
5CN0
 
 | | Artificial HIV fusion inhibitor AP2 fused to the C-terminus of gp41 NHR | | 分子名称: | DI(HYDROXYETHYL)ETHER, Envelope glycoprotein,AP2, MAGNESIUM ION | | 著者 | Zhu, Y, Ye, S, Zhang, R. | | 登録日 | 2015-07-17 | | 公開日 | 2015-09-16 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (1.901 Å) | | 主引用文献 | Improved Pharmacological and Structural Properties of HIV Fusion Inhibitor AP3 over Enfuvirtide: Highlighting Advantages of Artificial Peptide Strategy.
Sci Rep, 5, 2015
|
|
5CMU
 
 | |
8FA1
 
 | |
8FA2
 
 | |
4F7P
 
 | | Crystal Structure of HLA-A*2402 Complexed with a Newly Identified Peptide from 2009H1N1 PB1 (496-505) | | 分子名称: | Beta-2-microglobulin, HLA class I histocompatibility antigen, A-24 alpha chain, ... | | 著者 | Liu, J, Zhang, S, Tan, S, Yi, Y, Wu, B, Zhu, F, Wang, H, Qi, J, Gao, G.F. | | 登録日 | 2012-05-16 | | 公開日 | 2012-10-10 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Cross-Allele Cytotoxic T Lymphocyte Responses against 2009 Pandemic H1N1 Influenza A Virus among HLA-A24 and HLA-A3 Supertype-Positive Individuals.
J.Virol., 86, 2012
|
|
8GDS
 
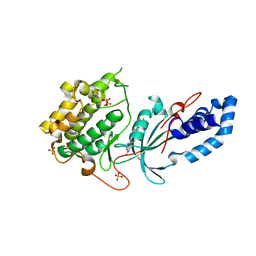 | |
6AIC
 
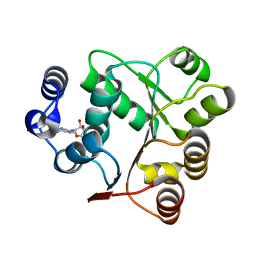 | | Crystal structures of the N-terminal domain of Staphylococcus aureus DEAD-box Cold shock RNA helicase CshA in complex with AMP | | 分子名称: | ADENOSINE MONOPHOSPHATE, DEAD-box ATP-dependent RNA helicase CshA | | 著者 | Tian, T, Chengliang, W, Xiaobao, C, Xuan, Z, Jianye, Z. | | 登録日 | 2018-08-22 | | 公開日 | 2018-11-21 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Crystal structures of the N-terminal domain of the Staphylococcus aureus DEAD-box RNA helicase CshA and its complex with AMP
Acta Crystallogr F Struct Biol Commun, 74, 2018
|
|
4F7M
 
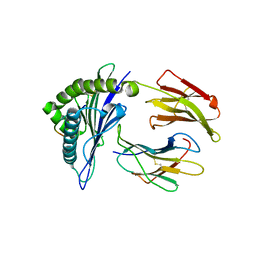 | | Crystal Structure of HLA-A*2402 Complexed with a Newly Identified Peptide from 2009 H1N1 PA (649-658) | | 分子名称: | Beta-2-microglobulin, HLA class I histocompatibility antigen, A-24 alpha chain, ... | | 著者 | Liu, J, Zhang, S, Tan, S, Yi, Y, Wu, B, Zhu, F, Wang, H, Qi, J, George, F.G. | | 登録日 | 2012-05-16 | | 公開日 | 2012-10-10 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Cross-Allele Cytotoxic T Lymphocyte Responses against 2009 Pandemic H1N1 Influenza A Virus among HLA-A24 and HLA-A3 Supertype-Positive Individuals.
J.Virol., 86, 2012
|
|
8GCQ
 
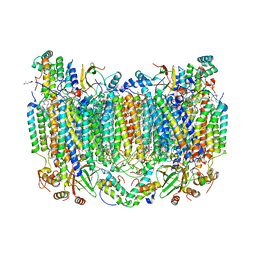 | | SFX structure of oxidized cytochrome c oxidase at 2.38 Angstrom resolution | | 分子名称: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(STEAROYLOXY)METHYL]ETHYL (5E,8E,11E,14E)-ICOSA-5,8,11,14-TETRAENOATE, (7R,17E,20E)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSA-17,20-DIEN-1-AMINIUM 4-OXIDE, ... | | 著者 | Ishigami, I, Yeh, S.-R, Rousseau, D.L. | | 登録日 | 2023-03-02 | | 公開日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.38 Å) | | 主引用文献 | Structural insights into functional properties of the oxidized form of cytochrome c oxidase.
Nat Commun, 14, 2023
|
|
5CVC
 
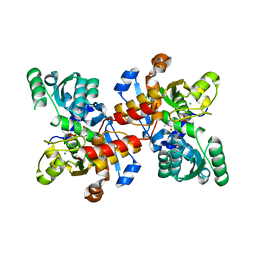 | | Structure of maize serine racemase | | 分子名称: | MAGNESIUM ION, PYRIDOXAL-5'-PHOSPHATE, Serine racemase | | 著者 | Song, Y, Zou, L, Fan, J. | | 登録日 | 2015-07-26 | | 公開日 | 2016-03-16 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.09 Å) | | 主引用文献 | Crystal structure of maize serine racemase with pyridoxal 5'-phosphate.
Acta Crystallogr.,Sect.F, 72, 2016
|
|
1KX9
 
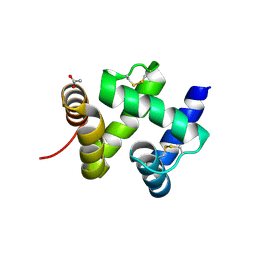 | | ANTENNAL CHEMOSENSORY PROTEIN A6 FROM THE MOTH MAMESTRA BRASSICAE | | 分子名称: | ACETATE ION, CHEMOSENSORY PROTEIN A6 | | 著者 | Lartigue, A, Campanacci, V, Roussel, A, Larsson, A.M, Jones, T.A, Tegoni, M, Cambillau, C. | | 登録日 | 2002-01-31 | | 公開日 | 2002-12-04 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | X-ray structure and ligand binding study of a moth chemosensory protein
J.Biol.Chem., 277, 2002
|
|
1KX8
 
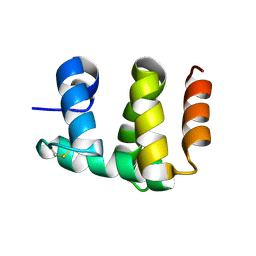 | | Antennal Chemosensory Protein A6 from Mamestra brassicae, tetragonal form | | 分子名称: | CHEMOSENSORY PROTEIN A6 | | 著者 | Lartigue, A, Campanacci, V, Roussel, A, Larsson, A.M, Jones, T.A, Tegoni, M, Cambillau, C. | | 登録日 | 2002-01-31 | | 公開日 | 2002-12-04 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | X-Ray Structure and Ligand Binding Study of a Chemosensory Protein
J.Biol.Chem., 277, 2002
|
|
6AIB
 
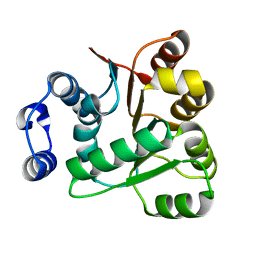 | | Crystal structures of the N-terminal RecA-like domain 1 of Staphylococcus aureus DEAD-box Cold shock RNA helicase CshA | | 分子名称: | DEAD-box ATP-dependent RNA helicase CshA | | 著者 | Chengliang, W, Tian, T, Xiaobao, C, Xuan, Z, Jianye, Z. | | 登録日 | 2018-08-22 | | 公開日 | 2018-11-21 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Crystal structures of the N-terminal domain of the Staphylococcus aureus DEAD-box RNA helicase CshA and its complex with AMP
Acta Crystallogr F Struct Biol Commun, 74, 2018
|
|
2JQB
 
 | | Solution structure of a novel D-amiNo acid containing conopeptide, conomarphin at pH 5 | | 分子名称: | Conomarphin | | 著者 | Huang, F, Du, W, Han, Y, Wang, C, Chi, C. | | 登録日 | 2007-05-31 | | 公開日 | 2008-06-03 | | 最終更新日 | 2023-12-20 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of a novel D-amiNo acid containing conopeptide, conomarphin at pH 5
To be Published
|
|
8HL4
 
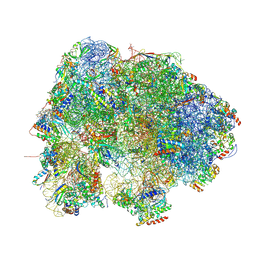 | |
4RFS
 
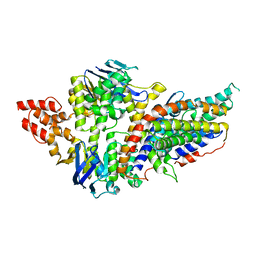 | | Structure of a pantothenate energy coupling factor transporter | | 分子名称: | Energy-coupling factor transporter ATP-binding protein EcfA1, Energy-coupling factor transporter ATP-binding protein EcfA2, Energy-coupling factor transporter transmembrane protein EcfT, ... | | 著者 | Zhang, M, Bao, Z, Zhao, Q, Guo, H, Xu, K, Zhang, P. | | 登録日 | 2014-09-27 | | 公開日 | 2014-12-24 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (3.232 Å) | | 主引用文献 | Structure of a pantothenate transporter and implications for ECF module sharing and energy coupling of group II ECF transporters.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
8HL1
 
 | |
2L01
 
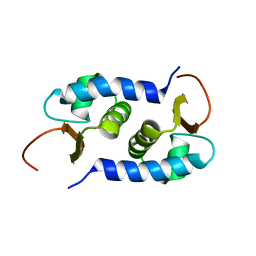 | | Solution NMR Structure of protein BVU3908 from Bacteroides vulgatus, Northeast Structural Genomics Consortium Target BvR153 | | 分子名称: | Uncharacterized protein | | 著者 | Eletsky, A, Lee, C, Wang, K, Ciccosanti, T.B, Hamilton, R, Acton, J.B, Xiao, G.B, Everett, J.K, Prestegard, J.H, Montelione, G.T, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | 登録日 | 2010-06-29 | | 公開日 | 2010-08-25 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution NMR Structure of protein BVU3908 from Bacteroides vulgatus
To be Published
|
|
