6BCD
 
 | | Crystal structure of Rev7-K44A/R124A/A135D in complex with Rev3-RBM2 (residues 1988-2014) | | 分子名称: | DNA polymerase zeta catalytic subunit, Mitotic spindle assembly checkpoint protein MAD2B | | 著者 | Rizzo, A.A, Hao, B, Li, Y, Korzhnev, D.M. | | 登録日 | 2017-10-20 | | 公開日 | 2018-08-01 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.43 Å) | | 主引用文献 | Rev7 dimerization is important for assembly and function of the Rev1/Pol zeta translesion synthesis complex.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6BC8
 
 | | Crystal structure of Rev7-R124A/Rev3-RBM2 (residues 1988-2014) complex | | 分子名称: | ACETATE ION, DNA polymerase zeta catalytic subunit, Mitotic spindle assembly checkpoint protein MAD2B, ... | | 著者 | Rizzo, A.A, Hao, B, Li, Y, Korzhnev, D.M. | | 登録日 | 2017-10-20 | | 公開日 | 2018-08-01 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.68 Å) | | 主引用文献 | Rev7 dimerization is important for assembly and function of the Rev1/Pol zeta translesion synthesis complex.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
7EQ4
 
 | | Crystal Structure of the N-terminus of Nonstructural protein 1 from SARS-CoV-2 | | 分子名称: | Host translation inhibitor nsp1 | | 著者 | Liu, Y, Ke, Z, Hu, H, Zhao, K, Xiao, J, Xia, Y, Li, Y. | | 登録日 | 2021-04-29 | | 公開日 | 2021-06-09 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.25 Å) | | 主引用文献 | Structural Basis and Function of the N Terminus of SARS-CoV-2 Nonstructural Protein 1.
Microbiol Spectr, 9, 2021
|
|
6BZE
 
 | | Cryo-EM structure of BCL10 CARD filament | | 分子名称: | B-cell lymphoma/leukemia 10 | | 著者 | David, L, Li, Y, Ma, J, Garner, E, Zhang, X, Wu, H. | | 登録日 | 2017-12-23 | | 公開日 | 2018-02-14 | | 最終更新日 | 2024-03-13 | | 実験手法 | ELECTRON MICROSCOPY (4 Å) | | 主引用文献 | Assembly mechanism of the CARMA1-BCL10-MALT1-TRAF6 signalosome.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6BI7
 
 | | Crystal structure of Rev7-WT/Rev3 as a monomer under high-salt conditions | | 分子名称: | DNA polymerase zeta catalytic subunit, Mitotic spindle assembly checkpoint protein MAD2B | | 著者 | Rizzo, A.A, Korzhnev, D.M, Hao, B, Li, Y. | | 登録日 | 2017-11-01 | | 公開日 | 2018-08-01 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Rev7 dimerization is important for assembly and function of the Rev1/Pol zeta translesion synthesis complex.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
7CZ0
 
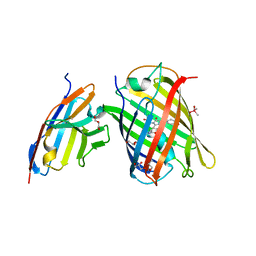 | | Crystal structure of a thermostable green fluorescent protein (TGP) with a synthetic nanobody (Sb92) | | 分子名称: | ACETATE ION, CACODYLATE ION, CACODYLIC ACID, ... | | 著者 | Cai, H, Yao, H, Li, T, Hutter, C, Tang, Y, Li, Y, Seeger, M, Li, D. | | 登録日 | 2020-09-06 | | 公開日 | 2021-09-08 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.77 Å) | | 主引用文献 | An improved fluorescent protein tag and its nanobodies for membrane protein expression, stability assay, and purification
To Be Published
|
|
6M49
 
 | | cryo-EM structure of Scap/Insig complex in the present of 25-hydroxyl cholesterol. | | 分子名称: | 25-HYDROXYCHOLESTEROL, Insulin-induced gene 2 protein, Sterol regulatory element-binding protein cleavage-activating protein,Sterol regulatory element-binding protein cleavage-activating protein | | 著者 | Yan, R, Cao, P, Song, W, Qian, H, Du, X, Coates, H.W, Zhao, X, Li, Y, Gao, S, Gong, X, Liu, X, Sui, J, Lei, J, Yang, H, Brown, A.J, Zhou, Q, Yan, C, Yan, N. | | 登録日 | 2020-03-06 | | 公開日 | 2021-01-20 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (3.7 Å) | | 主引用文献 | A structure of human Scap bound to Insig-2 suggests how their interaction is regulated by sterols.
Science, 371, 2021
|
|
5YFP
 
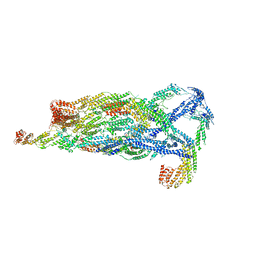 | | Cryo-EM Structure of the Exocyst Complex | | 分子名称: | Exocyst complex component EXO70, Exocyst complex component EXO84, Exocyst complex component SEC10, ... | | 著者 | Mei, K, Li, Y, Wang, S, Shao, G, Wang, J, Ding, Y, Luo, G, Yue, P, Liu, J.J, Wang, X, Dong, M.Q, Guo, W, Wang, H.W. | | 登録日 | 2017-09-21 | | 公開日 | 2018-01-31 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (4.4 Å) | | 主引用文献 | Cryo-EM structure of the exocyst complex
Nat. Struct. Mol. Biol., 25, 2018
|
|
6M7A
 
 | |
5Y26
 
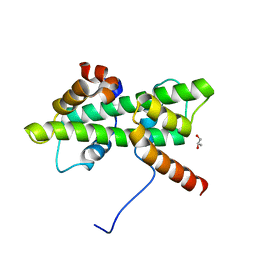 | | Crystal structure of native Dpb4-Dpb3 | | 分子名称: | DNA polymerase epsilon subunit D, GLYCEROL, Putative transcription factor C16C4.22 | | 著者 | Chen, Y.H, Li, Y, Gao, F. | | 登録日 | 2017-07-24 | | 公開日 | 2018-01-24 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (2.003 Å) | | 主引用文献 | Coordinated regulation of heterochromatin inheritance by Dpb3-Dpb4 complex.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5Y44
 
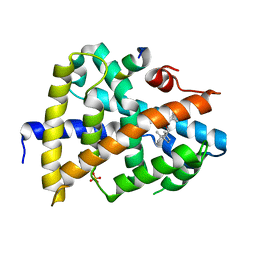 | | A novel moderator XD4 for bile acid receptor | | 分子名称: | Bile acid receptor, Peptide from Nuclear receptor coactivator 1, SULFATE ION, ... | | 著者 | Lu, Y, Li, Y. | | 登録日 | 2017-08-01 | | 公開日 | 2019-02-20 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (2.35 Å) | | 主引用文献 | A novel moderator XD4 for bile acid receptor
To Be Published
|
|
6M7B
 
 | |
6MEW
 
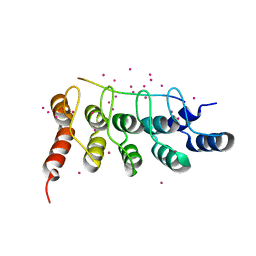 | | RFXANK ankyrin repeats in complex with a RFX7 peptide | | 分子名称: | DNA-binding protein RFXANK, RFX7 peptide, UNKNOWN ATOM OR ION | | 著者 | Tempel, W, Xu, C, Dong, A, Li, Y, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | 登録日 | 2018-09-07 | | 公開日 | 2018-10-03 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.78 Å) | | 主引用文献 | RFXANK ankyrin repeats in complex with a RFX7 peptide
to be published
|
|
5Y1J
 
 | |
5VSM
 
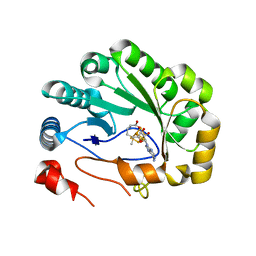 | | Crystal structure of viperin with bound [4Fe-4S] cluster, 5'-deoxyadenosine, and L-methionine | | 分子名称: | 5'-DEOXYADENOSINE, IRON/SULFUR CLUSTER, METHIONINE, ... | | 著者 | Fenwick, M.K, Li, Y, Cresswell, P, Modis, Y, Ealick, S.E. | | 登録日 | 2017-05-11 | | 公開日 | 2017-06-14 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Structural studies of viperin, an antiviral radical SAM enzyme.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5Y49
 
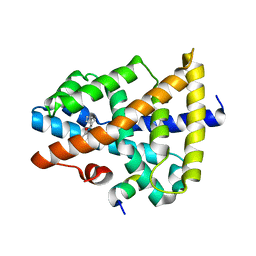 | | A moderator XD22 binding to bile acid receptor | | 分子名称: | Bile acid receptor, Peptide from Nuclear receptor coactivator 2, [(1R,2S,4S)-2-bicyclo[2.2.1]heptanyl] 4-azanylbenzoate | | 著者 | Lu, Y, Li, Y. | | 登録日 | 2017-08-02 | | 公開日 | 2019-02-20 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | A moderator XD22 binding to bile acid receptor
To Be Published
|
|
6N31
 
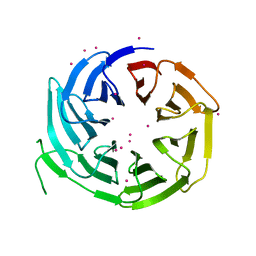 | | WD repeats of human WDR12 | | 分子名称: | Ribosome biogenesis protein WDR12, UNKNOWN ATOM OR ION | | 著者 | Halabelian, L, Zeng, H, Tempel, W, Li, Y, Seitova, A, Hutchinson, A, Bountra, C, Edwards, A.M, Arrowsmith, C.H, Structural Genomics Consortium (SGC) | | 登録日 | 2018-11-14 | | 公開日 | 2018-12-12 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | WD repeats of human WDR12
To Be Published
|
|
6MRW
 
 | | 14-meric ClyA pore complex | | 分子名称: | Hemolysin E, chromosomal | | 著者 | Peng, W, de Souza Santos, M, Li, Y, Tomchick, D.R, Orth, K. | | 登録日 | 2018-10-15 | | 公開日 | 2019-05-15 | | 最終更新日 | 2024-03-13 | | 実験手法 | ELECTRON MICROSCOPY (4.3 Å) | | 主引用文献 | High-resolution cryo-EM structures of the E. coli hemolysin ClyA oligomers.
Plos One, 14, 2019
|
|
6MRT
 
 | | 12-meric ClyA pore complex | | 分子名称: | Hemolysin E, chromosomal | | 著者 | Peng, W, de Souza Santos, M, Li, Y, Tomchick, D.R, Orth, K. | | 登録日 | 2018-10-15 | | 公開日 | 2019-05-15 | | 最終更新日 | 2024-03-13 | | 実験手法 | ELECTRON MICROSCOPY (2.8 Å) | | 主引用文献 | High-resolution cryo-EM structures of the E. coli hemolysin ClyA oligomers.
Plos One, 14, 2019
|
|
6MB2
 
 | |
6NBS
 
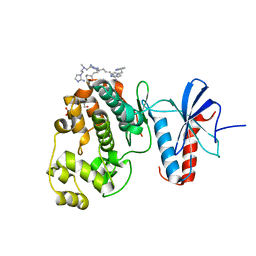 | | WT ERK2 with compound 2507-8 | | 分子名称: | (5S)-5-benzyl-4,5-dihydro-1H-imidazol-2-amine, GLYCEROL, Mitogen-activated protein kinase 1, ... | | 著者 | Sammons, R.M, Perry, N.A, Cho, E.J, Kaoud, T.S, Zamora-Olivares, D.P, Piserchio, A, Houghten, R.A, Giulianotti, M, Li, Y, Debevec, G, Gurevich, V.V, Ghose, R, Iverson, T.M, Dalby, K.N. | | 登録日 | 2018-12-10 | | 公開日 | 2019-07-31 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | A Novel Class of Common Docking Domain Inhibitors That Prevent ERK2 Activation and Substrate Phosphorylation.
Acs Chem.Biol., 14, 2019
|
|
6MRU
 
 | | 13-meric ClyA pore complex | | 分子名称: | Hemolysin E, chromosomal | | 著者 | Peng, W, de Souza Santos, M, Li, Y, Tomchick, D.R, Orth, K. | | 登録日 | 2018-10-15 | | 公開日 | 2019-05-15 | | 最終更新日 | 2024-03-13 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | High-resolution cryo-EM structures of the E. coli hemolysin ClyA oligomers.
Plos One, 14, 2019
|
|
5VSL
 
 | | Crystal structure of viperin with bound [4Fe-4S] cluster and S-adenosylhomocysteine (SAH) | | 分子名称: | IRON/SULFUR CLUSTER, Radical S-adenosyl methionine domain-containing protein 2, S-ADENOSYL-L-HOMOCYSTEINE | | 著者 | Fenwick, M.K, Li, Y, Cresswell, P, Modis, Y, Ealick, S.E. | | 登録日 | 2017-05-11 | | 公開日 | 2017-06-14 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (1.972 Å) | | 主引用文献 | Structural studies of viperin, an antiviral radical SAM enzyme.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
7XMK
 
 | | Crystal structure of human RIPK1 kinase domain in complex with compound SKLB923 | | 分子名称: | 5-[2-(cyclopropylcarbonylamino)-[1,2,4]triazolo[1,5-a]pyridin-7-yl]-N-[(1S)-1-(3-fluorophenyl)ethyl]-1-methyl-indole-3-carboxamide, IODIDE ION, Receptor-interacting serine/threonine-protein kinase 1 | | 著者 | Zhang, L, Wang, Y, Li, Y, Yang, S. | | 登録日 | 2022-04-26 | | 公開日 | 2023-04-26 | | 最終更新日 | 2024-05-15 | | 実験手法 | X-RAY DIFFRACTION (2.376 Å) | | 主引用文献 | From Hit to Lead: Structure-Based Optimization of Novel Selective Inhibitors of Receptor-Interacting Protein Kinase 1 (RIPK1) for the Treatment of Inflammatory Diseases.
J.Med.Chem., 67, 2024
|
|
8SZV
 
 | | Crystal structure of pregnane X receptor ligand binding domain complexed with T0901317 analog SJPYT-318 | | 分子名称: | N-[4-(1,1,1,3,3,3-hexafluoro-2-hydroxypropan-2-yl)phenyl]-N-[(4-phenoxyphenyl)methyl]benzenesulfonamide, Nuclear receptor subfamily 1 group I member 2 | | 著者 | Huber, A.D, Poudel, S, Miller, D.J, Li, Y, Chen, T. | | 登録日 | 2023-05-30 | | 公開日 | 2023-10-11 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Ligand flexibility and binding pocket malleability cooperate to allow selective PXR activation by analogs of a promiscuous nuclear receptor ligand.
Structure, 31, 2023
|
|
