1A5Y
 
 | |
7K4X
 
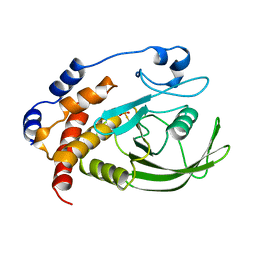
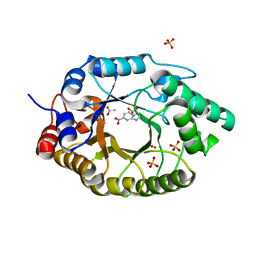 | | Crystal structure of Kemp Eliminase HG3.7 in complex with the transition state analog 6-nitrobenzotriazole | | 分子名称: | 6-NITROBENZOTRIAZOLE, ACETATE ION, Endo-1,4-beta-xylanase, ... | | 著者 | Padua, R.A.P, Otten, R, Bunzel, A, Nguyen, V, Pitsawong, W, Patterson, M, Sui, S, Perry, S.L, Cohen, A.E, Hilvert, D, Kern, D. | | 登録日 | 2020-09-16 | | 公開日 | 2020-12-02 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | How directed evolution reshapes the energy landscape in an enzyme to boost catalysis.
Science, 370, 2020
|
|
5EUE
 
 | | S1P Lyase Bacterial Surrogate bound to N-(2-((4-methoxy-2,5-dimethylbenzyl)amino)-1-phenylethyl)-5-methylisoxazole-3-carboxamide | | 分子名称: | PHOSPHATE ION, Putative sphingosine-1-phosphate lyase, ~{N}-[(1~{S})-2-[(4-methoxy-2,5-dimethyl-phenyl)methylamino]-1-phenyl-ethyl]-5-methyl-1,2-oxazole-3-carboxamide | | 著者 | Argiriadi, M.A, Banach, D, Radziejewska, E, Marchie, S, DiMauro, J, Dinges, J, Dominguez, E, Hutchins, C, Judge, R.A, Queeney, K, Wallace, G, Harris, C.M. | | 登録日 | 2015-11-18 | | 公開日 | 2016-03-16 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.83 Å) | | 主引用文献 | Creation of a S1P Lyase bacterial surrogate for structure-based drug design.
Bioorg.Med.Chem.Lett., 26, 2016
|
|
5EUD
 
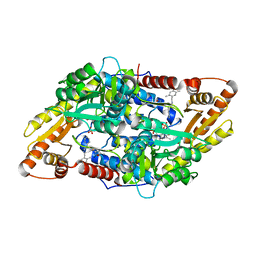
 | | S1P Lyase Bacterial Surrogate bound to N-(1-(4-(3-hydroxyprop-1-yn-1-yl)phenyl)-2-((4-methoxy-2,5-dimethylbenzyl)amino)ethyl)-5-methylisoxazole-3-carboxamide | | 分子名称: | PHOSPHATE ION, Putative sphingosine-1-phosphate lyase, ~{N}-[(1~{S})-2-[(4-methoxy-2,5-dimethyl-phenyl)methylamino]-1-[4-(3-oxidanylprop-1-ynyl)phenyl]ethyl]-5-methyl-1,2-oxazole-3-carboxamide | | 著者 | Argiriadi, M.A, Banach, D, Radziejewska, E, Marchie, S, DiMauro, J, Dinges, J, Dominguez, E, Hutchins, C, Judge, R.A, Queeney, K, Wallace, G, Harris, C.M. | | 登録日 | 2015-11-18 | | 公開日 | 2016-03-16 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.24 Å) | | 主引用文献 | Creation of a S1P Lyase bacterial surrogate for structure-based drug design.
Bioorg.Med.Chem.Lett., 26, 2016
|
|
8R6U
 
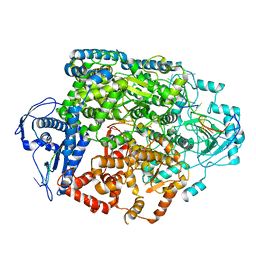 | | Structure of the SFTSV L protein in a transcription-priming state without capped RNA [TRANSCRIPTION-PRIMING (in vitro)] | | 分子名称: | MAGNESIUM ION, RNA (5'-R(P*CP*UP*GP*GP*GP*CP*GP*GP*UP*CP*UP*UP*UP*GP*UP*GP*U)-3'), RNA primer, ... | | 著者 | Williams, H.M, Thorkelsson, S.R, Vogel, D, Busch, C, Milewski, M, Cusack, S, Grunewald, K, Quemin, E.R.J, Rosenthal, M. | | 登録日 | 2023-11-23 | | 公開日 | 2024-04-24 | | 最終更新日 | 2024-06-19 | | 実験手法 | ELECTRON MICROSCOPY (2.98 Å) | | 主引用文献 | Structural snapshots of phenuivirus cap-snatching and transcription.
Nucleic Acids Res., 52, 2024
|
|
8R6Y
 
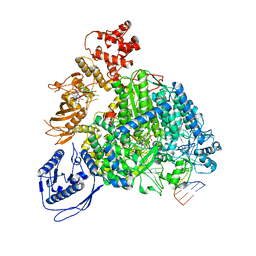 | | Structure of the SFTSV L protein stalled in a transcription-specific early elongation state with bound capped RNA [TRANSCRIPTION-EARLY-ELONGATION] | | 分子名称: | 5'-O-[(S)-hydroxy{[(S)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]uridine, MAGNESIUM ION, RNA (5'-R(*(M7G)*AP*AP*A)-3'), ... | | 著者 | Williams, H.M, Thorkelsson, S.R, Vogel, D, Busch, C, Milewski, M, Cusack, S, Grunewald, K, Quemin, E.R.J, Rosenthal, M. | | 登録日 | 2023-11-23 | | 公開日 | 2024-04-24 | | 最終更新日 | 2024-06-19 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | Structural snapshots of phenuivirus cap-snatching and transcription.
Nucleic Acids Res., 52, 2024
|
|
8R6W
 
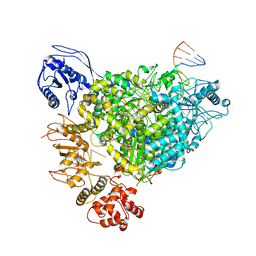 | | Structure of the SFTSV L protein in a transcription-priming state with bound capped RNA [TRANSCRIPTION-PRIMING] | | 分子名称: | RNA (5'-R(*(M7G)*AP*AP*A)-3'), RNA (5'-R(*AP*CP*AP*C)-3'), RNA (5'-R(*AP*CP*AP*CP*AP*GP*AP*GP*AP*CP*GP*CP*CP*CP*AP*G)-3'), ... | | 著者 | Williams, H.M, Thorkelsson, S.R, Vogel, D, Busch, C, Milewski, M, Cusack, S, Grunewald, K, Quemin, E.R.J, Rosenthal, M. | | 登録日 | 2023-11-23 | | 公開日 | 2024-04-24 | | 最終更新日 | 2024-06-19 | | 実験手法 | ELECTRON MICROSCOPY (3.35 Å) | | 主引用文献 | Structural snapshots of phenuivirus cap-snatching and transcription.
Nucleic Acids Res., 52, 2024
|
|
7OJH
 
 | | ABCG2 topotecan turnover-1 state | | 分子名称: | (S)-10-[(DIMETHYLAMINO)METHYL]-4-ETHYL-4,9-DIHYDROXY-1H-PYRANO[3',4':6,7]INOLIZINO[1,2-B]-QUINOLINE-3,14(4H,12H)-DIONE, ADENOSINE-5'-TRIPHOSPHATE, Broad substrate specificity ATP-binding cassette transporter ABCG2, ... | | 著者 | Yu, Q, Ni, D, Kowal, J, Manolaridis, I, Jackson, S.M, Stahlberg, H, Locher, K.P. | | 登録日 | 2021-05-15 | | 公開日 | 2021-07-21 | | 最終更新日 | 2021-07-28 | | 実験手法 | ELECTRON MICROSCOPY (3.1 Å) | | 主引用文献 | Structures of ABCG2 under turnover conditions reveal a key step in the drug transport mechanism.
Nat Commun, 12, 2021
|
|
7K4Z
 
 | | Crystal structure of Kemp Eliminase HG3.17 in complex with the transition state analog 6-nitrobenzotriazole | | 分子名称: | 6-NITROBENZOTRIAZOLE, Endo-1,4-beta-xylanase, PENTAETHYLENE GLYCOL | | 著者 | Padua, R.A.P, Otten, R, Bunzel, A, Nguyen, V, Pitsawong, W, Patterson, M, Sui, S, Perry, S.L, Cohen, A.E, Hilvert, D, Kern, D. | | 登録日 | 2020-09-16 | | 公開日 | 2020-12-02 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.08 Å) | | 主引用文献 | How directed evolution reshapes the energy landscape in an enzyme to boost catalysis.
Science, 370, 2020
|
|
7K4Q
 
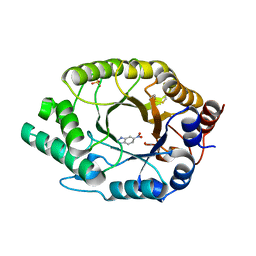 | | Crystal structure of Kemp Eliminase HG3 in complex with the transition state analog 6-nitrobenzotriazole | | 分子名称: | 6-NITROBENZOTRIAZOLE, Endo-1,4-beta-xylanase, SULFATE ION | | 著者 | Padua, R.A.P, Otten, R, Bunzel, A, Nguyen, V, Pitsawong, W, Patterson, M, Sui, S, Perry, S.L, Cohen, A.E, Hilvert, D, Kern, D. | | 登録日 | 2020-09-16 | | 公開日 | 2020-12-02 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.28 Å) | | 主引用文献 | How directed evolution reshapes the energy landscape in an enzyme to boost catalysis.
Science, 370, 2020
|
|
7K4V
 
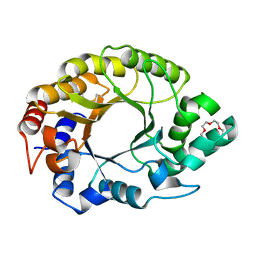 | | Crystal structure of Kemp Eliminase HG3.17 | | 分子名称: | Endo-1,4-beta-xylanase, TETRAETHYLENE GLYCOL | | 著者 | Padua, R.A.P, Otten, R, Bunzel, A, Nguyen, V, Pitsawong, W, Patterson, M, Sui, S, Perry, S.L, Cohen, A.E, Hilvert, D, Kern, D. | | 登録日 | 2020-09-16 | | 公開日 | 2020-12-02 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.3 Å) | | 主引用文献 | How directed evolution reshapes the energy landscape in an enzyme to boost catalysis.
Science, 370, 2020
|
|
5ETX
 
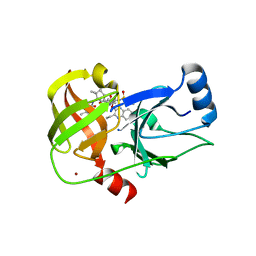 | | Crystal structure of HCV NS3/4A protease A156T variant in complex with 5172-Linear (MK-5172 linear analogue) | | 分子名称: | CHLORIDE ION, NS3 protease, ZINC ION, ... | | 著者 | Soumana, D, Yilmaz, N.K, Ali, A, Prachanronarong, K.L, Aydin, C, Schiffer, C.A. | | 登録日 | 2015-11-18 | | 公開日 | 2016-01-13 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.35 Å) | | 主引用文献 | Structural and Thermodynamic Effects of Macrocyclization in HCV NS3/4A Inhibitor MK-5172.
Acs Chem.Biol., 11, 2016
|
|
4QX1
 
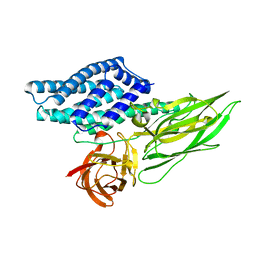 | | Cry3A Toxin structure obtained by Serial Femtosecond Crystallography from in vivo grown crystals isolated from Bacillus thuringiensis and data processed with the CrystFEL software suite | | 分子名称: | Pesticidal crystal protein cry3Aa | | 著者 | Sawaya, M.R, Cascio, D, Gingery, M, Rodriguez, J, Goldschmidt, L, Colletier, J.-P, Messerschmidt, M, Boutet, S, Koglin, J.E, Williams, G.J, Brewster, A.S, Nass, K, Hattne, J, Botha, S, Doak, R.B, Shoeman, R.L, DePonte, D.P, Park, H.-W, Federici, B.A, Sauter, N.K, Schlichting, I, Eisenberg, D. | | 登録日 | 2014-07-17 | | 公開日 | 2014-08-13 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Protein crystal structure obtained at 2.9 angstrom resolution from injecting bacterial cells into an X-ray free-electron laser beam.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
5GVR
 
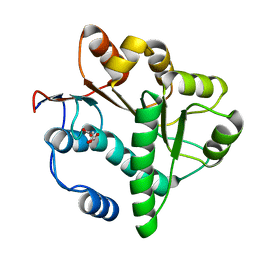 | | Crystal structure of the DDX41 DEAD domain in an apo closed form | | 分子名称: | (2S)-2-hydroxybutanedioic acid, Probable ATP-dependent RNA helicase DDX41 | | 著者 | Omura, H, Oikawa, D, Nakane, T, Kato, M, Ishii, R, Goto, Y, Suga, H, Ishitani, R, Tokunaga, F, Nureki, O. | | 登録日 | 2016-09-06 | | 公開日 | 2016-10-19 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Structural and Functional Analysis of DDX41: a bispecific immune receptor for DNA and cyclic dinucleotide
Sci Rep, 6, 2016
|
|
3OOC
 
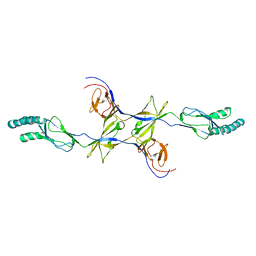 | | Crystal structure of the membrane fusion protein CusB from Escherichia coli | | 分子名称: | Cation efflux system protein cusB | | 著者 | Su, C.-C, Yang, F, Long, F, Reyon, D, Routh, M.D, Kuo, D.W, Mokhtari, A.K, Van Ornam, J.D, Rabe, K.L, Hoy, J.A, Lee, Y.J, Rajashankar, K.R, Yu, E.W. | | 登録日 | 2010-08-30 | | 公開日 | 2010-12-29 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (3.404 Å) | | 主引用文献 | Crystal structure of the membrane fusion protein CusB from Escherichia coli.
J.Mol.Biol., 393, 2009
|
|
5GXT
 
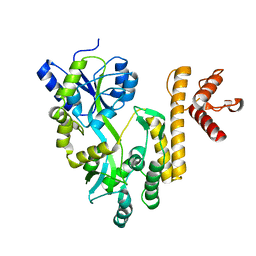 | | Crystal structure of PigG | | 分子名称: | MAGNESIUM ION, Maltose-binding periplasmic protein,PigG | | 著者 | Zhang, F, Ran, T, Xu, D, Wang, W. | | 登録日 | 2016-09-20 | | 公開日 | 2017-07-19 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.245 Å) | | 主引用文献 | Crystal structure of MBP-PigG fusion protein and the essential function of PigG in the prodigiosin biosynthetic pathway in Serratia marcescens FS14.
Int. J. Biol. Macromol., 99, 2017
|
|
3OTL
 
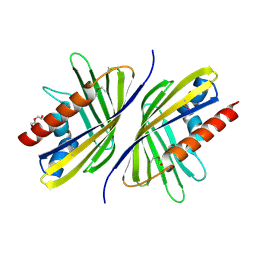 | | Three-dimensional Structure of the putative uncharacterized protein from Rhizobium leguminosarum at the resolution 1.9A, Northeast Structural Genomics Consortium Target RlR261 | | 分子名称: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, DI(HYDROXYETHYL)ETHER, Putative uncharacterized protein | | 著者 | Kuzin, A, Lew, S, Seetharaman, J, Mao, M, Xiao, R, Ciccosanti, C, Wang, D, Everett, J.K, Nair, R, Acton, T.B, Rost, B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | 登録日 | 2010-09-13 | | 公開日 | 2010-09-22 | | 最終更新日 | 2021-10-06 | | 実験手法 | X-RAY DIFFRACTION (1.901 Å) | | 主引用文献 | Northeast Structural Genomics Consortium Target RlR261
To be Published
|
|
7OJ8
 
 | | ABCG2 E1S turnover-2 state | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, Broad substrate specificity ATP-binding cassette transporter ABCG2, CHOLESTEROL, ... | | 著者 | Ni, D, Yu, Q, Kowal, J, Manolaridis, I, Jackson, S.M, Stahlberg, H, Locher, K.P. | | 登録日 | 2021-05-14 | | 公開日 | 2021-07-21 | | 最終更新日 | 2021-07-28 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | Structures of ABCG2 under turnover conditions reveal a key step in the drug transport mechanism.
Nat Commun, 12, 2021
|
|
7OJI
 
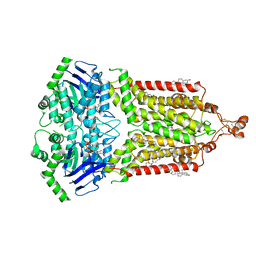 | | ABCG2 topotecan turnover-2 state | | 分子名称: | (S)-10-[(DIMETHYLAMINO)METHYL]-4-ETHYL-4,9-DIHYDROXY-1H-PYRANO[3',4':6,7]INOLIZINO[1,2-B]-QUINOLINE-3,14(4H,12H)-DIONE, ADENOSINE-5'-TRIPHOSPHATE, Broad substrate specificity ATP-binding cassette transporter ABCG2, ... | | 著者 | Yu, Q, Ni, D, Kowal, J, Manolaridis, I, Jackson, S.M, Stahlberg, H, Locher, K.P. | | 登録日 | 2021-05-16 | | 公開日 | 2021-07-21 | | 最終更新日 | 2021-07-28 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | Structures of ABCG2 under turnover conditions reveal a key step in the drug transport mechanism.
Nat Commun, 12, 2021
|
|
3SPA
 
 | | Crystal Structure of Human Mitochondrial RNA Polymerase | | 分子名称: | CHLORIDE ION, DNA-directed RNA polymerase, mitochondrial, ... | | 著者 | Ringel, R, Sologub, M, Morozov, Y.I, Litonin, D, Cramer, P, Temiakov, D. | | 登録日 | 2011-07-01 | | 公開日 | 2011-09-28 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Structure of human mitochondrial RNA polymerase
Nature, 478, 2011
|
|
7P5C
 
 | | Cryo-EM structure of human TTYH3 in Ca2+ and GDN | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Protein tweety homolog 3 | | 著者 | Sukalskaia, A, Straub, M.S, Sawicka, M, Deneka, D, Dutzler, R. | | 登録日 | 2021-07-14 | | 公開日 | 2021-08-11 | | 最終更新日 | 2021-09-08 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Cryo-EM structures of the TTYH family reveal a novel architecture for lipid interactions.
Nat Commun, 12, 2021
|
|
5GRJ
 
 | | Crystal structure of human PD-L1 with monoclonal antibody avelumab | | 分子名称: | Programmed cell death 1 ligand 1, avelumab H chain, avelumab L chain | | 著者 | Liu, K, Tan, S, Chai, Y, Chen, D, Song, H, Zhang, C.W.-H, Shi, Y, Liu, J, Tan, W, Lyu, J, Gao, S, Yan, J, Qi, J, Gao, G.F. | | 登録日 | 2016-08-11 | | 公開日 | 2016-11-02 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (3.206 Å) | | 主引用文献 | Structural basis of anti-PD-L1 monoclonal antibody avelumab for tumor therapy.
Cell Res., 27, 2017
|
|
5GTT
 
 | | Crystal structure of C. perfringens iota-like enterotoxin CPILE-a | | 分子名称: | 1,2-ETHANEDIOL, Binary enterotoxin of Clostridium perfringens component a | | 著者 | Toniti, W, Yoshida, T, Tsurumura, T, Irikura, D, Tsuge, H. | | 登録日 | 2016-08-23 | | 公開日 | 2017-03-01 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.011 Å) | | 主引用文献 | Crystal structure and structure-based mutagenesis of actin-specific ADP-ribosylating toxin CPILE-a as novel enterotoxin
PLoS ONE, 12, 2017
|
|
8QOH
 
 | | Crystal structure of the kinetoplastid kinetochore protein KKT14 C-terminal domain from Apiculatamorpha spiralis | | 分子名称: | kinetochore protein KKT14 | | 著者 | Carter, W, Ballmer, D, Ishii, M, Ludzia, P, Akiyoshi, B. | | 登録日 | 2023-09-28 | | 公開日 | 2024-01-17 | | 最終更新日 | 2024-06-19 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Kinetoplastid kinetochore proteins KKT14-KKT15 are divergent Bub1/BubR1-Bub3 proteins.
Open Biology, 14, 2024
|
|
5H1P
 
 | | CRISPR-associated protein | | 分子名称: | ACETATE ION, CRISPR-associated endoribonuclease Cas2 | | 著者 | Ka, D, Jeong, U, Bae, E. | | 登録日 | 2016-10-11 | | 公開日 | 2017-10-18 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Structural and dynamic insights into the role of conformational switching in the nuclease activity of the Xanthomonas albilineans Cas2 in CRISPR-mediated adaptive immunity
Struct Dyn, 4, 2017
|
|
