1F40
 
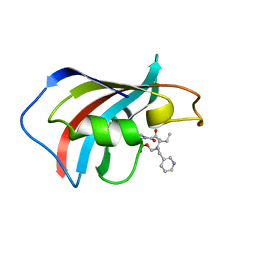 | | SOLUTION STRUCTURE OF FKBP12 COMPLEXED WITH GPI-1046, A NEUROTROPHIC LIGAND | | 分子名称: | (2S)-[3-PYRIDYL-1-PROPYL]-1-[3,3-DIMETHYL-1,2-DIOXOPENTYL]-2-PYRROLIDINECARBOXYLATE, FK506 BINDING PROTEIN (FKBP12) | | 著者 | Sich, C, Improta, S, Cowley, D.J, Guenet, C, Merly, J.P, Teufel, M, Saudek, V. | | 登録日 | 2000-06-07 | | 公開日 | 2000-11-08 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of a neurotrophic ligand bound to FKBP12 and its effects on protein dynamics.
Eur.J.Biochem., 267, 2000
|
|
1F4H
 
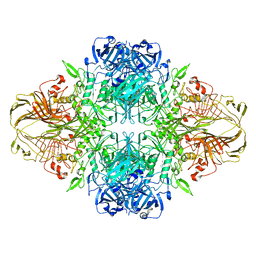 | | E. COLI (LACZ) BETA-GALACTOSIDASE (ORTHORHOMBIC) | | 分子名称: | BETA-GALACTOSIDASE, MAGNESIUM ION | | 著者 | Juers, D.H, Jacobson, R.H, Wigley, D, Zhang, X.J, Huber, R.E, Tronrud, D.E, Matthews, B.W. | | 登録日 | 2000-06-07 | | 公開日 | 2001-02-21 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | High resolution refinement of beta-galactosidase in a new crystal form reveals multiple metal-binding sites and provides a structural basis for alpha-complementation.
Protein Sci., 9, 2000
|
|
1F4A
 
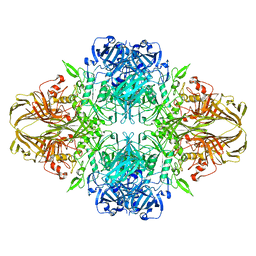 | | E. COLI (LACZ) BETA-GALACTOSIDASE (NCS CONSTRAINED MONOMER-ORTHORHOMBIC) | | 分子名称: | BETA-GALACTOSIDASE, MAGNESIUM ION | | 著者 | Juers, D.H, Jacobson, R.H, Wigley, D, Zhang, X.J, Huber, R.E, Tronrud, D.E, Matthews, B.W. | | 登録日 | 2000-06-07 | | 公開日 | 2001-02-21 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | High resolution refinement of beta-galactosidase in a new crystal form reveals multiple metal-binding sites and provides a structural basis for alpha-complementation.
Protein Sci., 9, 2000
|
|
1FKI
 
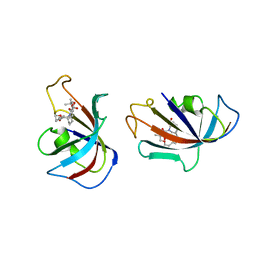 | | DESIGN, SYNTHESIS, AND KINETIC EVALUATION OF HIGH-AFFINITY FKBP LIGANDS, AND THE X-RAY CRYSTAL STRUCTURES OF THEIR COMPLEXES WITH FKBP12 | | 分子名称: | (21S)-1AZA-4,4-DIMETHYL-6,19-DIOXA-2,3,7,20-TETRAOXOBICYCLO[19.4.0] PENTACOSANE, FK506 BINDING PROTEIN | | 著者 | Holt, D.A, Luengo, J.I, Yamashita, D.S, Oh, H.-J, Konialian, A.L, Yen, H.-K, Rozamus, L.W, Brandt, M, Bossard, M.J, Levy, M.A, Eggleston, D.S, Stout, T.J, Liang, J, Schultz, L.W, Clardy, J. | | 登録日 | 1993-08-05 | | 公開日 | 1994-01-31 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | DESIGN, SYNTHESIS, AND KINETIC EVALUATION OF HIGH-AFFINITY FKBP LIGANDS AND THE X-RAY CRYSTAL-STRUCTURES OF THEIR COMPLEXES WITH FKBP12.
J.Am.Chem.Soc., 115, 1993
|
|
1FKR
 
 | | SOLUTION STRUCTURE OF FKBP, A ROTAMASE ENZYME AND RECEPTOR FOR FK506 AND RAPAMYCIN | | 分子名称: | FK506 AND RAPAMYCIN-BINDING PROTEIN | | 著者 | Michnick, S.W, Rosen, M.K, Wandless, T.J, Karplus, M, Schreiber, S.L. | | 登録日 | 1992-03-05 | | 公開日 | 1994-01-31 | | 最終更新日 | 2024-05-01 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of FKBP, a rotamase enzyme and receptor for FK506 and rapamycin.
Science, 252, 1991
|
|
1FKT
 
 | | SOLUTION STRUCTURE OF FKBP, A ROTAMASE ENZYME AND RECEPTOR FOR FK506 AND RAPAMYCIN | | 分子名称: | FK506 AND RAPAMYCIN-BINDING PROTEIN | | 著者 | Michnick, S.W, Rosen, M.K, Wandless, T.J, Karplus, M, Schreiber, S.L. | | 登録日 | 1992-03-05 | | 公開日 | 1994-01-31 | | 最終更新日 | 2024-05-01 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of FKBP, a rotamase enzyme and receptor for FK506 and rapamycin.
Science, 252, 1991
|
|
1FKG
 
 | | DESIGN, SYNTHESIS, AND KINETIC EVALUATION OF HIGH-AFFINITY FKBP LIGANDS, AND THE X-RAY CRYSTAL STRUCTURES OF THEIR COMPLEXES WITH FKBP12 | | 分子名称: | 1,3-DIPHENYL-1-PROPYL-1-(3,3-DIMETHYL-1,2-DIOXYPENTYL)-2-PIPERIDINE CARBOXYLATE, FK506 BINDING PROTEIN | | 著者 | Holt, D.A, Luengo, J.I, Yamashita, D.S, Oh, H.-J, Konialian, A.L, Yen, H.-K, Rozamus, L.W, Brandt, M, Bossard, M.J, Levy, M.A, Eggleston, D.S, Stout, T.J, Liang, J, Schultz, L.W, Clardy, J. | | 登録日 | 1993-08-05 | | 公開日 | 1994-01-31 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | DESIGN, SYNTHESIS, AND KINETIC EVALUATION OF HIGH-AFFINITY FKBP LIGANDS AND THE X-RAY CRYSTAL-STRUCTURES OF THEIR COMPLEXES WITH FKBP12.
J.Am.Chem.Soc., 115, 1993
|
|
1BLC
 
 | |
1BMP
 
 | | BONE MORPHOGENETIC PROTEIN-7 | | 分子名称: | BONE MORPHOGENETIC PROTEIN-7 | | 著者 | Griffith, D.L, Scott, D.L. | | 登録日 | 1995-12-14 | | 公開日 | 1997-07-23 | | 最終更新日 | 2024-11-20 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Three-dimensional structure of recombinant human osteogenic protein 1: structural paradigm for the transforming growth factor beta superfamily.
Proc.Natl.Acad.Sci.USA, 93, 1996
|
|
1D6O
 
 | | NATIVE FKBP | | 分子名称: | AMMONIUM ION, PROTEIN (FK506-BINDING PROTEIN), SULFATE ION | | 著者 | Burkhard, P, Taylor, P, Walkinshaw, M.D. | | 登録日 | 1999-10-15 | | 公開日 | 1999-10-21 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | X-ray structures of small ligand-FKBP complexes provide an estimate for hydrophobic interaction energies.
J.Mol.Biol., 295, 2000
|
|
1FKS
 
 | | SOLUTION STRUCTURE OF FKBP, A ROTAMASE ENZYME AND RECEPTOR FOR FK506 AND RAPAMYCIN | | 分子名称: | FK506 AND RAPAMYCIN-BINDING PROTEIN | | 著者 | Michnick, S.W, Rosen, M.K, Wandless, T.J, Karplus, M, Schreiber, S.L. | | 登録日 | 1992-03-05 | | 公開日 | 1994-01-31 | | 最終更新日 | 2024-05-01 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of FKBP, a rotamase enzyme and receptor for FK506 and rapamycin.
Science, 252, 1991
|
|
1FRT
 
 | |
1FKH
 
 | | DESIGN, SYNTHESIS, AND KINETIC EVALUATION OF HIGH-AFFINITY FKBP LIGANDS, AND THE X-RAY CRYSTAL STRUCTURES OF THEIR COMPLEXES WITH FKBP12 | | 分子名称: | 1-CYCLOHEXYL-3-PHENYL-1-PROPYL-1-(3,3-DIMETHYL-1,2-DIOXYPENTYL)-2-PIPERIDINE CARBOXYLATE, FK506 BINDING PROTEIN | | 著者 | Holt, D.A, Luengo, J.I, Yamashita, D.S, Oh, H.-J, Konialian, A.L, Yen, H.-K, Rozamus, L.W, Brandt, M, Bossard, M.J, Levy, M.A, Eggleston, D.S, Stout, T.J, Liang, J, Schultz, L.W, Clardy, J. | | 登録日 | 1993-08-05 | | 公開日 | 1994-01-31 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | DESIGN, SYNTHESIS, AND KINETIC EVALUATION OF HIGH-AFFINITY FKBP LIGANDS AND THE X-RAY CRYSTAL-STRUCTURES OF THEIR COMPLEXES WITH FKBP12.
J.Am.Chem.Soc., 115, 1993
|
|
1DAT
 
 | | CUBIC CRYSTAL STRUCTURE RECOMBINANT HORSE L APOFERRITIN | | 分子名称: | CADMIUM ION, L FERRITIN | | 著者 | Gallois, B, Granier, T, Langlois D'Estaintot, B, Crichton, R.R, Roland, F. | | 登録日 | 1996-11-14 | | 公開日 | 1997-03-12 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (2.05 Å) | | 主引用文献 | X-ray structure of recombinant horse L-chain apoferritin at 2.0 angstrom resolution: Implications for stability and function.
J.Biol.Inorg.Chem., 2, 1997
|
|
1EMY
 
 | |
8D8R
 
 | | SARS-CoV-2 Spike RBD in complex with DMAb 2196 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2196 heavy chain, 2196 light chain, ... | | 著者 | Du, J, Cui, J, Pallesen, J. | | 登録日 | 2022-06-08 | | 公開日 | 2022-10-19 | | 最終更新日 | 2024-11-06 | | 実験手法 | ELECTRON MICROSCOPY (4.1 Å) | | 主引用文献 | DNA-delivered antibody cocktail exhibits improved pharmacokinetics and confers prophylactic protection against SARS-CoV-2.
Nat Commun, 13, 2022
|
|
8D8Q
 
 | | SARS-CoV-2 Spike RBD in complex with DMAbs 2130 and 2196 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2130 heavy chain, 2130 light chain, ... | | 著者 | Du, J, Cui, J, Pallesen, J. | | 登録日 | 2022-06-08 | | 公開日 | 2022-10-19 | | 最終更新日 | 2024-10-23 | | 実験手法 | ELECTRON MICROSCOPY (4.2 Å) | | 主引用文献 | DNA-delivered antibody cocktail exhibits improved pharmacokinetics and confers prophylactic protection against SARS-CoV-2.
Nat Commun, 13, 2022
|
|
8UF6
 
 | | Structure of Trek-1(K2P2.1) with ML336 | | 分子名称: | CADMIUM ION, DECANE, DODECANE, ... | | 著者 | Lolicato, M, Mondal, A, Minor, D.L. | | 登録日 | 2023-10-03 | | 公開日 | 2024-06-26 | | 最終更新日 | 2025-01-08 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Development of covalent chemogenetic K 2P channel activators.
Cell Chem Biol, 31, 2024
|
|
8UEC
 
 | | Structure of TREK-1CG*:CAT335a | | 分子名称: | CADMIUM ION, DECANE, HEXADECANE, ... | | 著者 | Mondal, A, Lee, H, Minor, D.L. | | 登録日 | 2023-09-30 | | 公開日 | 2024-06-26 | | 最終更新日 | 2025-01-08 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Development of covalent chemogenetic K 2P channel activators.
Cell Chem Biol, 31, 2024
|
|
8UE2
 
 | | Structure of TREK-1CG*:ML335 | | 分子名称: | CADMIUM ION, DECANE, HEXADECANE, ... | | 著者 | Mondal, A, Lee, H, Minor, D.L. | | 登録日 | 2023-09-29 | | 公開日 | 2024-06-26 | | 最終更新日 | 2025-01-08 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Development of covalent chemogenetic K 2P channel activators.
Cell Chem Biol, 31, 2024
|
|
8UE9
 
 | | Structure of TREK-1CG*:CAT335 | | 分子名称: | CADMIUM ION, HEXADECANE, N-((E,2S,3R)-1,3-DIHYDROXYOCTADEC-4-EN-2-YL)PALMITAMIDE, ... | | 著者 | Mondal, A, Lee, H, Minor, D.L. | | 登録日 | 2023-09-29 | | 公開日 | 2024-06-26 | | 最終更新日 | 2025-01-08 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Development of covalent chemogenetic K 2P channel activators.
Cell Chem Biol, 31, 2024
|
|
9BWF
 
 | | Crystal structure of cellulose oxidative enzyme without ligand | | 分子名称: | COPPER (II) ION, Cellulose oxidative enzyme | | 著者 | Morais, M.A.B, Santos, C.A, Araujo, E.A, Santos, C.R, Morao, L.G, Motta, M.L, Murakami, M.T. | | 登録日 | 2024-05-21 | | 公開日 | 2024-12-04 | | 最終更新日 | 2025-04-02 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | A metagenomic 'dark matter' enzyme catalyses oxidative cellulose conversion.
Nature, 639, 2025
|
|
9BWH
 
 | | Crystal structure of cellulose oxidative enzyme with glycerol | | 分子名称: | COPPER (II) ION, Cellulose oxidative enzyme, GLYCEROL | | 著者 | Morais, M.A.B, Santos, C.A, Araujo, E.A, Santos, C.R, Morao, L.G, Motta, M.L, Murakami, M.T. | | 登録日 | 2024-05-21 | | 公開日 | 2024-12-04 | | 最終更新日 | 2025-04-02 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | A metagenomic 'dark matter' enzyme catalyses oxidative cellulose conversion.
Nature, 639, 2025
|
|
9BWI
 
 | | Crystal structure of cellulose oxidative enzyme in acidic pH with glycerol | | 分子名称: | COPPER (II) ION, Cellulose oxidative enzyme, GLYCEROL | | 著者 | Morais, M.A.B, Santos, C.A, Araujo, E.A, Santos, C.R, Morao, L.G, Motta, M.L, Murakami, M.T. | | 登録日 | 2024-05-21 | | 公開日 | 2024-12-11 | | 最終更新日 | 2025-04-02 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | A metagenomic 'dark matter' enzyme catalyses oxidative cellulose conversion.
Nature, 639, 2025
|
|
8F5Y
 
 | | Crystal structure of pregnane X receptor ligand binding domain complexed with JQ1 | | 分子名称: | (6S)-6-(2-tert-butoxy-2-oxoethyl)-4-(4-chlorophenyl)-2,3,9-trimethyl-6,7-dihydrothieno[3,2-f][1,2,4]triazolo[4,3-a][1,4]diazepin-10-ium, Nuclear receptor coactivator 1, Nuclear receptor subfamily 1 group I member 2 | | 著者 | Huber, A.D, Poudel, S, Seetharaman, J, Miller, D.J, Chen, T. | | 登録日 | 2022-11-15 | | 公開日 | 2024-02-21 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (2.15 Å) | | 主引用文献 | A bromodomain-independent mechanism of gene regulation by the BET inhibitor JQ1: direct activation of nuclear receptor PXR.
Nucleic Acids Res., 52, 2024
|
|
