6D2Q
 
 | |
5K9N
 
 | | Structural and Mechanistic Analysis of Drosophila melanogaster Polyamine N acetyltransferase, an enzyme that Catalyzes the Formation of N acetylagmatine | | 分子名称: | Polyamine N acetyltransferase | | 著者 | Dempsey, D.R, Nichols, D.A, Battistini, M.R, Pemberton, O, Ospina, S.R, Zhang, X, Carpenter, A.-M, Chen, Y, Merkler, D.J. | | 登録日 | 2016-06-01 | | 公開日 | 2017-06-07 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Structural and Mechanistic Analysis of Drosophila melanogaster Agmatine N-Acetyltransferase, an Enzyme that Catalyzes the Formation of N-Acetylagmatine.
Sci Rep, 7, 2017
|
|
6D21
 
 | |
5YCO
 
 | | Complex structure of PCNA with UHRF2 | | 分子名称: | E3 ubiquitin-protein ligase UHRF2, GLYCEROL, Proliferating cell nuclear antigen, ... | | 著者 | Wu, M, Chen, W, Hang, T, Wang, C, Zhang, X, Zang, J. | | 登録日 | 2017-09-07 | | 公開日 | 2017-11-15 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.199 Å) | | 主引用文献 | Structure insights into the molecular mechanism of the interaction between UHRF2 and PCNA.
Biochem. Biophys. Res. Commun., 494, 2017
|
|
2LIT
 
 | | NMR Solution Structure of Yeast Iso-1-cytochrome c Mutant P71H in reduced states | | 分子名称: | Cytochrome c iso-1, HEME C | | 著者 | Lan, W, Wang, Z, Yang, Z, Zhu, J, Ying, T, Jiang, X, Zhang, X, Wu, H, Liu, M, Tan, X, Cao, C, Huang, Z.X. | | 登録日 | 2011-08-31 | | 公開日 | 2011-12-07 | | 最終更新日 | 2024-10-16 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Conformational toggling of yeast iso-1-cytochrome C in the oxidized and reduced States.
Plos One, 6, 2011
|
|
3AIY
 
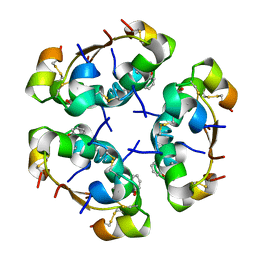 | | R6 HUMAN INSULIN HEXAMER (SYMMETRIC), NMR, REFINED AVERAGE STRUCTURE | | 分子名称: | PHENOL, PROTEIN (INSULIN) | | 著者 | O'Donoghue, S.I, Chang, X, Abseher, R, Nilges, M, Led, J.J. | | 登録日 | 1998-12-29 | | 公開日 | 2000-02-28 | | 最終更新日 | 2024-10-30 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Unraveling the symmetry ambiguity in a hexamer: calculation of the R6 human insulin structure.
J.Biomol.NMR, 16, 2000
|
|
7XTZ
 
 | | Structure of SARS-CoV-2 Spike Protein with Engineered x3 Disulfide (x3(D427C, V987C) and single Arg S1/S2 cleavage site), Locked-1 Conformation | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BILIVERDINE IX ALPHA, ... | | 著者 | Qu, K, Chen, Q, Ciazynska, K.A, Liu, B, Zhang, X, Wang, J, He, Y, Guan, J, He, J, Liu, T, Zhang, X, Carter, A.P, Xiong, X, Briggs, J.A.G. | | 登録日 | 2022-05-18 | | 公開日 | 2022-07-20 | | 最終更新日 | 2025-06-25 | | 実験手法 | ELECTRON MICROSCOPY (2.8 Å) | | 主引用文献 | Engineered disulfide reveals structural dynamics of locked SARS-CoV-2 spike.
Plos Pathog., 18, 2022
|
|
3LNL
 
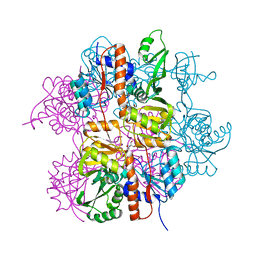 | | Crystal structure of Staphylococcus aureus protein SA1388 | | 分子名称: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, UPF0135 protein SA1388, ZINC ION | | 著者 | Singh, K.S, Chruszcz, M, Zhang, X, Minor, W, Zhang, H. | | 登録日 | 2010-02-02 | | 公開日 | 2010-03-16 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structure of a Conserved Hypothetical Protein Sa1388 from S. aureus Reveals a Capped Hexameric Toroid with Two Pii Domain Lids and a Dinuclear Metal Center.
Bmc Struct.Biol., 6, 2006
|
|
7XU1
 
 | | Structure of SARS-CoV-2 Spike Protein with Engineered x3 Disulfide (x3(D427C, V987C) and single Arg S1/S2 cleavage site), Locked-122 Conformation | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BILIVERDINE IX ALPHA, ... | | 著者 | Qu, K, Chen, Q, Ciazynska, K.A, Liu, B, Zhang, X, Wang, J, He, Y, Guan, J, He, J, Liu, T, Zhang, X, Carter, A.P, Xiong, X, Briggs, J.A.G. | | 登録日 | 2022-05-18 | | 公開日 | 2022-07-20 | | 最終更新日 | 2025-06-25 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | Engineered disulfide reveals structural dynamics of locked SARS-CoV-2 spike.
Plos Pathog., 18, 2022
|
|
7XU4
 
 | | Structure of SARS-CoV-2 D614G Spike Protein with Engineered x3 Disulfide (x3(D427C, V987C) and single Arg S1/S2 cleavage site), Locked-2 Conformation | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BILIVERDINE IX ALPHA, ... | | 著者 | Qu, K, Chen, Q, Ciazynska, K.A, Liu, B, Zhang, X, Wang, J, He, Y, Guan, J, He, J, Liu, T, Zhang, X, Carter, A.P, Xiong, X, Briggs, J.A.G. | | 登録日 | 2022-05-18 | | 公開日 | 2022-07-20 | | 最終更新日 | 2025-07-02 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Engineered disulfide reveals structural dynamics of locked SARS-CoV-2 spike.
Plos Pathog., 18, 2022
|
|
7XU5
 
 | | Structure of SARS-CoV-2 D614G Spike Protein with Engineered x3 Disulfide (x3(D427C, V987C) and single Arg S1/S2 cleavage site), Closed Conformation | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BILIVERDINE IX ALPHA, ... | | 著者 | Qu, K, Chen, Q, Ciazynska, K.A, Liu, B, Zhang, X, Wang, J, He, Y, Guan, J, He, J, Liu, T, Zhang, X, Carter, A.P, Xiong, X, Briggs, J.A.G. | | 登録日 | 2022-05-18 | | 公開日 | 2022-07-20 | | 最終更新日 | 2025-06-18 | | 実験手法 | ELECTRON MICROSCOPY (3.1 Å) | | 主引用文献 | Engineered disulfide reveals structural dynamics of locked SARS-CoV-2 spike.
Plos Pathog., 18, 2022
|
|
7XU3
 
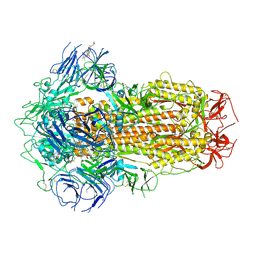 | | Structure of SARS-CoV-2 Spike Protein with Engineered x3 Disulfide (x3(D427C, V987C) and single Arg S1/S2 cleavage site), Closed Conformation | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BILIVERDINE IX ALPHA, ... | | 著者 | Qu, K, Chen, Q, Ciazynska, K.A, Liu, B, Zhang, X, Wang, J, He, Y, Guan, J, He, J, Liu, T, Zhang, X, Carter, A.P, Xiong, X, Briggs, J.A.G. | | 登録日 | 2022-05-18 | | 公開日 | 2022-07-20 | | 最終更新日 | 2025-06-18 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | Engineered disulfide reveals structural dynamics of locked SARS-CoV-2 spike.
Plos Pathog., 18, 2022
|
|
7XU6
 
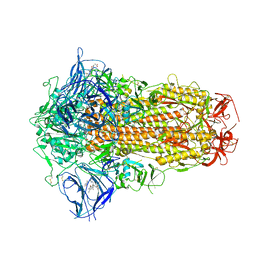 | | Structure of SARS-CoV-2 Spike Protein with Engineered x3 Disulfide (x3(D427C, V987C) and single Arg S1/S2 cleavage site), incubated in Low pH after 40-Day Storage in PBS, Locked-2 Conformation | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BILIVERDINE IX ALPHA, ... | | 著者 | Qu, K, Chen, Q, Ciazynska, K.A, Liu, B, Zhang, X, Wang, J, He, Y, Guan, J, He, J, Liu, T, Zhang, X, Carter, A.P, Xiong, X, Briggs, J.A.G. | | 登録日 | 2022-05-18 | | 公開日 | 2022-07-20 | | 最終更新日 | 2025-07-02 | | 実験手法 | ELECTRON MICROSCOPY (2.9 Å) | | 主引用文献 | Engineered disulfide reveals structural dynamics of locked SARS-CoV-2 spike.
Plos Pathog., 18, 2022
|
|
9IK4
 
 | |
3LZZ
 
 | | Crystal structures of Cupin superfamily BbDUF985 from Branchiostoma belcheri tsingtauense in apo and GDP-bound forms | | 分子名称: | ACETATE ION, GUANOSINE-5'-DIPHOSPHATE, Putative uncharacterized protein | | 著者 | Du, Y, He, Y.-X, Saren, G, Zhang, X, Zhang, S.-C, Chen, Y, Zhou, C.-Z. | | 登録日 | 2010-03-02 | | 公開日 | 2010-06-02 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Crystal structures of the apo and GDP-bound forms of a cupin-like protein BbDUF985 from Branchiostoma belcheri tsingtauense
Proteins, 2010
|
|
8F49
 
 | |
8F7Y
 
 | |
8EW2
 
 | |
8EW0
 
 | |
3LOI
 
 | | Crystal structures of Cupin superfamily BbDUF985 from Branchiostoma belcheri tsingtauense in the apo and GDP-bound forms | | 分子名称: | Putative uncharacterized protein | | 著者 | Zou, C.Z, Du, Y, He, Y.-X, Saren, G, Zhang, X, Chen, Y, Zhang, S.-C. | | 登録日 | 2010-02-04 | | 公開日 | 2010-06-02 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Crystal structures of the apo and GDP-bound forms of a cupin-like protein BbDUF985 from Branchiostoma belcheri tsingtauense
Proteins, 2010
|
|
8EXE
 
 | |
8EXF
 
 | |
9JF8
 
 | |
4LT5
 
 | | Structure of a Naegleria Tet-like dioxygenase in complex with 5-methylcytosine DNA | | 分子名称: | 1,2-ETHANEDIOL, DNA, MANGANESE (II) ION, ... | | 著者 | Hashimoto, H, Pais, J.E, Zhang, X, Saleh, L, Fu, Z.Q, Dai, N, Correa, I.R, Roberts, R.J, Zheng, Y, Cheng, X. | | 登録日 | 2013-07-23 | | 公開日 | 2013-12-18 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.893 Å) | | 主引用文献 | Structure of a Naegleria Tet-like dioxygenase in complex with 5-methylcytosine DNA.
Nature, 506, 2014
|
|
4M9V
 
 | | Zfp57 mutant (E182Q) in complex with 5-carboxylcytosine DNA | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, ACETATE ION, CALCIUM ION, ... | | 著者 | Liu, Y, Olanrewaju, Y.O, Zhang, X, Cheng, X. | | 登録日 | 2013-08-15 | | 公開日 | 2013-11-27 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (0.969 Å) | | 主引用文献 | DNA recognition of 5-carboxylcytosine by a zfp57 mutant at an atomic resolution of 0.97 angstrom.
Biochemistry, 52, 2013
|
|
