1R4V
 
 | | 1.9A crystal structure of protein AQ328 from Aquifex aeolicus | | 分子名称: | CACODYLATE ION, Hypothetical protein AQ_328, ZINC ION | | 著者 | Qiu, Y, Tereshko, V, Kim, Y, Zhang, R, Collart, F, Joachimiak, A, Kossiakoff, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2003-10-08 | | 公開日 | 2004-03-30 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | The crystal structure of Aq_328 from the hyperthermophilic bacteria Aquifex aeolicus shows an ancestral histone fold.
Proteins, 62, 2006
|
|
1S6Y
 
 | | 2.3A crystal structure of phospho-beta-glucosidase | | 分子名称: | 6-phospho-beta-glucosidase | | 著者 | Tereshko, V, Dementieva, I, Kim, Y, Collat, F, Joachimiak, A, Kossiakoff, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2004-01-28 | | 公開日 | 2004-05-25 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (2.31 Å) | | 主引用文献 | 2.3A CRYSTAL STRUCTURE OF PHOSPHO-BETA-GLUCOSIDASE, licH Gene Product from BACILLUS STEAROTHERMOPHILUS
To be Published
|
|
4ZQN
 
 | | Crystal Structure of the Catalytic Domain of the Inosine Monophosphate Dehydrogenase from Mycobacterium tuberculosis in the complex with IMP and the inhibitor P41 | | 分子名称: | 2-chloro-N,N-dimethyl-5-[({2-[3-(prop-1-en-2-yl)phenyl]propan-2-yl}carbamoyl)amino]benzamide, INOSINIC ACID, Inosine-5'-monophosphate dehydrogenase,Inosine-5'-monophosphate dehydrogenase | | 著者 | Kim, Y, Makowska-Grzyska, M, Gu, M, Kavitha, M, Hedstrom, L, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2015-05-10 | | 公開日 | 2015-06-17 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Mycobacterium tuberculosis IMPDH in Complexes with Substrates, Products and Antitubercular Compounds.
Plos One, 10, 2015
|
|
2IQY
 
 | | Rat Phosphatidylethanolamine-Binding Protein | | 分子名称: | CALCIUM ION, CHLORIDE ION, Phosphatidylethanolamine-binding protein 1 | | 著者 | Kim, Y, Joachimiak, G, Heil, G.L, Koide, S, Joachimiak, A. | | 登録日 | 2006-10-14 | | 公開日 | 2007-09-25 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | Crystal Structure of Rat Phosphatidylethanolamine-Binding Protein
To be Published
|
|
4P98
 
 | |
4ZDN
 
 | | Streptomyces platensis isomigrastatin ketosynthase domain MgsF KS4 | | 分子名称: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, AT-less polyketide synthase, CHLORIDE ION | | 著者 | Chang, C, Li, H, Endres, M, Bingman, C.A, Yennamalli, R, Lohman, J.R, Ma, M, Shen, B, Phillips Jr, G.N, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | 登録日 | 2015-04-17 | | 公開日 | 2015-05-13 | | 最終更新日 | 2023-03-22 | | 実験手法 | X-RAY DIFFRACTION (2.509 Å) | | 主引用文献 | Structural and evolutionary relationships of "AT-less" type I polyketide synthase ketosynthases.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
1U14
 
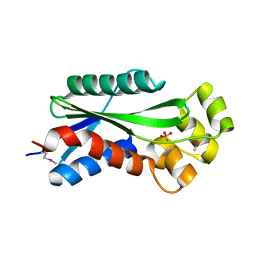 | | The crystal structure of hypothetical UPF0244 protein yjjX at resolution 1.68 Angstrom | | 分子名称: | Hypothetical UPF0244 protein yjjX, PHOSPHATE ION | | 著者 | Qiu, Y, Kim, Y, Cuff, M, Collart, F, Joachimiak, A, Kossiakoff, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2004-07-14 | | 公開日 | 2004-09-21 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (1.68 Å) | | 主引用文献 | The crystal structure of hypothetical UPF0244 protein yjjX at resolution 1.68 Angstrom
To be Published
|
|
4ZQM
 
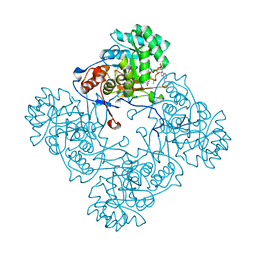 | | Crystal Structure of the Catalytic Domain of the Inosine Monophosphate Dehydrogenase from Mycobacterium tuberculosis in the complex with XMP and NAD | | 分子名称: | Inosine-5'-monophosphate dehydrogenase,Inosine-5'-monophosphate dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, XANTHOSINE-5'-MONOPHOSPHATE | | 著者 | Kim, Y, Maltseva, N, Makowska-Grzyska, M, Gu, M, Kavitha, M, Hedstrom, L, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2015-05-10 | | 公開日 | 2015-06-17 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (1.602 Å) | | 主引用文献 | Mycobacterium tuberculosis IMPDH in Complexes with Substrates, Products and Antitubercular Compounds.
Plos One, 10, 2015
|
|
4Z5P
 
 | | Crystal structure of the LnmA cytochrome P450 hydroxylase from the leinamycin biosynthetic pathway of Streptomyces atroolivaceus S-140 at 1.9 A resolution | | 分子名称: | Cytochrome P450 hydroxylase, PROTOPORPHYRIN IX CONTAINING FE, TRIETHYLENE GLYCOL | | 著者 | Ma, M, Lohman, J, Rudolf, J, Miller, M.D, Cao, H, Osipiuk, J, Babnigg, G, Phillips Jr, G.N, Joachimiak, A, Shen, B, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | 登録日 | 2015-04-02 | | 公開日 | 2015-07-29 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Crystal structure of the LnmA cytochrome P450 hydroxylase from the leinamycin biosynthetic pathway of Streptomyces atroolivaceus S-140
To be Published
|
|
4ZO4
 
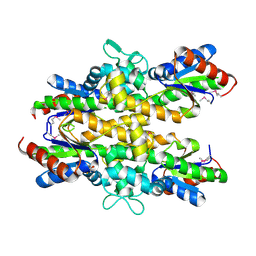 | | Dephospho-CoA kinase from Campylobacter jejuni. | | 分子名称: | BETA-MERCAPTOETHANOL, Dephospho-CoA kinase | | 著者 | Osipiuk, J, Zhou, M, Stam, J, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2015-05-06 | | 公開日 | 2015-05-13 | | 最終更新日 | 2019-12-11 | | 実験手法 | X-RAY DIFFRACTION (2.57 Å) | | 主引用文献 | Dephospho-CoA kinase from Campylobacter jejuni.
to be published
|
|
4ZPJ
 
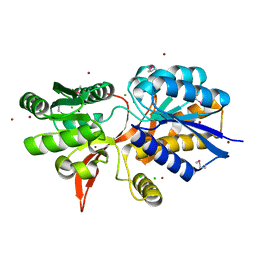 | | ABC transporter substrate-binding protein from Sphaerobacter thermophilus | | 分子名称: | CHLORIDE ION, Extracellular ligand-binding receptor, ZINC ION | | 著者 | OSIPIUK, J, Holowicki, J, Clancy, S, JOACHIMIAK, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2015-05-07 | | 公開日 | 2015-05-20 | | 最終更新日 | 2019-12-25 | | 実験手法 | X-RAY DIFFRACTION (2.24 Å) | | 主引用文献 | ABC transporter substrate-binding protein from Sphaerobacter thermophilus.
to be published
|
|
4YYF
 
 | | The crystal structure of a glycosyl hydrolase of GH3 family member from [Mycobacterium smegmatis str. MC2 155 | | 分子名称: | ACETATE ION, Beta-N-acetylhexosaminidase, FORMIC ACID, ... | | 著者 | Tan, K, Hatzos-Skintges, C, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2015-03-23 | | 公開日 | 2015-04-08 | | 最終更新日 | 2019-12-25 | | 実験手法 | X-RAY DIFFRACTION (1.92 Å) | | 主引用文献 | The crystal structure of a glycosyl hydrolase of GH3 family member from [Mycobacterium smegmatis str. MC2 155
To Be Published
|
|
4ZR7
 
 | | The structure of a domain of a functionally unknown protein from Bacillus subtilis subsp. subtilis str. 168 | | 分子名称: | ACETATE ION, CHLORIDE ION, Sensor histidine kinase ResE | | 著者 | Tan, K, Li, H, Jedrzejczak, R, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2015-05-11 | | 公開日 | 2015-05-27 | | 最終更新日 | 2019-12-25 | | 実験手法 | X-RAY DIFFRACTION (1.86 Å) | | 主引用文献 | The structure of a domain of a functionally unknown protein from Bacillus subtilis subsp. subtilis str. 168
To Be Published
|
|
4ZQP
 
 | | Crystal Structure of the Catalytic Domain of the Inosine Monophosphate Dehydrogenase from Mycobacterium tuberculosis in the complex with IMP and the inhibitor MAD1 | | 分子名称: | 5'-O-({1-[(2E)-4-(4-hydroxy-6-methoxy-7-methyl-3-oxo-1,3-dihydro-2-benzofuran-5-yl)-2-methylbut-2-en-1-yl]-1H-1,2,3-triazol-4-yl}methyl)adenosine, GLYCEROL, INOSINIC ACID, ... | | 著者 | Kim, Y, Makowska-Grzyska, M, Gu, M, Kavitha, M, Hedstrom, L, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2015-05-10 | | 公開日 | 2015-06-17 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Mycobacterium tuberculosis IMPDH in Complexes with Substrates, Products and Antitubercular Compounds.
Plos One, 10, 2015
|
|
4ZNM
 
 | | Crystal structure of SgcC5 protein from Streptomyces globisporus (apo form) | | 分子名称: | C-domain type II peptide synthetase, CHLORIDE ION, SODIUM ION | | 著者 | Michalska, K, Bigelow, L, Jedrzejczak, R, Babnigg, G, Lohman, J, Ma, M, Rudolf, J, Chang, C.-Y, Shen, B, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | 登録日 | 2015-05-04 | | 公開日 | 2015-05-27 | | 最終更新日 | 2019-12-25 | | 実験手法 | X-RAY DIFFRACTION (1.998 Å) | | 主引用文献 | Crystal structure of SgcC5 protein from Streptomyces globisporus (apo form)
To Be Published
|
|
4ZQR
 
 | | Crystal Structure of the Catalytic Domain of the Inosine Monophosphate Dehydrogenase from Mycobacterium tuberculosis | | 分子名称: | GLYCEROL, Inosine-5'-monophosphate dehydrogenase,Inosine-5'-monophosphate dehydrogenase, PHOSPHATE ION, ... | | 著者 | Kim, Y, Makowska-Grzyska, M, Gu, M, Kavitha, M, Hedstrom, L, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2015-05-11 | | 公開日 | 2015-06-17 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (1.692 Å) | | 主引用文献 | Mycobacterium tuberculosis IMPDH in Complexes with Substrates, Products and Antitubercular Compounds.
Plos One, 10, 2015
|
|
4ZV9
 
 | | 2.00 Angstrom resolution crystal structure of an uncharacterized protein from Escherichia coli O157:H7 str. Sakai | | 分子名称: | DI(HYDROXYETHYL)ETHER, GLYCEROL, PHOSPHATE ION, ... | | 著者 | Halavaty, A.S, Wawrzak, Z, Filippova, E.V, Kiryukhina, O, Endres, M, Joachimiak, A, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2015-05-18 | | 公開日 | 2015-06-17 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | 2.00 Angstrom resolution crystal structure of an uncharacterized protein from Escherichia coli O157:H7 str. Sakai
To Be Published
|
|
4ZWV
 
 | | Crystal Structure of Aminotransferase AtmS13 from Actinomadura melliaura | | 分子名称: | GLYCEROL, Putative aminotransferase | | 著者 | Kim, Y, Bigelow, L, Endres, M, Wang, F, Phillips Jr, G.N, Joachimiak, A, Enzyme Discovery for Natural Product Biosynthesis (NatPro), Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2015-05-19 | | 公開日 | 2015-06-03 | | 最終更新日 | 2019-12-04 | | 実験手法 | X-RAY DIFFRACTION (1.503 Å) | | 主引用文献 | Structural characterization of AtmS13, a putative sugar aminotransferase involved in indolocarbazole AT2433 aminopentose biosynthesis.
Proteins, 83, 2015
|
|
1S3X
 
 | | The crystal structure of the human Hsp70 ATPase domain | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, CALCIUM ION, Heat shock 70 kDa protein 1, ... | | 著者 | Sriram, M, Osipiuk, J, Freeman, B, Morimoto, R.I, Joachimiak, A. | | 登録日 | 2004-01-14 | | 公開日 | 2004-01-20 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (1.84 Å) | | 主引用文献 | Human Hsp70 molecular chaperone binds two calcium ions within the ATPase domain
Structure, 5, 1997
|
|
1S9U
 
 | | Atomic structure of a putative anaerobic dehydrogenase component | | 分子名称: | DI(HYDROXYETHYL)ETHER, SULFATE ION, putative component of anaerobic dehydrogenases | | 著者 | Qiu, Y, Zhang, R, Tereshko, V, Kim, Y, Collart, F, Joachimiak, A, Kossiakoff, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2004-02-05 | | 公開日 | 2004-06-08 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (1.38 Å) | | 主引用文献 | The 1.38 A crystal structure of DmsD protein from Salmonella typhimurium, a proofreading chaperone on the Tat pathway.
Proteins, 71, 2008
|
|
4MPT
 
 | | Crystal Structure of Periplasmic binding Protein Type 1 from Bordetella pertussis Tohama I | | 分子名称: | ACETIC ACID, Putative leu/ile/val-binding protein, SODIUM ION | | 著者 | Kim, Y, Joachimiak, G, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2013-09-13 | | 公開日 | 2013-12-11 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Crystal Structure of Periplasmic binding Protein Type 1 from Bordetella pertussis Tohama I
To be Published
|
|
4ZQO
 
 | | Crystal Structure of the Catalytic Domain of the Inosine Monophosphate Dehydrogenase from Mycobacterium tuberculosis in the complex with IMP and the inhibitor Q67 | | 分子名称: | GLYCEROL, INOSINIC ACID, Inosine-5'-monophosphate dehydrogenase,Inosine-5'-monophosphate dehydrogenase, ... | | 著者 | Kim, Y, Makowska-Grzyska, M, Gu, M, Kavitha, M, Hedstrom, L, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2015-05-10 | | 公開日 | 2015-06-17 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (1.76 Å) | | 主引用文献 | Mycobacterium tuberculosis IMPDH in Complexes with Substrates, Products and Antitubercular Compounds.
Plos One, 10, 2015
|
|
4Z5Q
 
 | | Crystal structure of the LnmZ cytochrome P450 hydroxylase from the leinamycin biosynthetic pathway of Streptomyces atroolivaceus S-140 at 1.8 A resolution | | 分子名称: | 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, CHLORIDE ION, Cytochrome P450 hydroxylase, ... | | 著者 | Ma, M, Lohman, J, Rudolf, J, Miller, M.D, Cao, H, Osipiuk, J, Joachimiak, A, Phillips Jr, G.N, Shen, B, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | 登録日 | 2015-04-02 | | 公開日 | 2015-08-12 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.801 Å) | | 主引用文献 | Crystal structure of the LnmZ cytochrome P450 hydroxylase from the leinamycin biosynthetic pathway of Streptomyces atroolivaceus S-140
To be Published
|
|
4ZHB
 
 | | N-terminal structure of ankyrin repeat-containing protein legA11 from Legionella pneumophila | | 分子名称: | 5-mer peptide, ACETATE ION, Ankyrin repeat-containing protein | | 著者 | Chang, C, Endres, M, Mack, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2015-04-24 | | 公開日 | 2015-05-06 | | 実験手法 | X-RAY DIFFRACTION (1.3 Å) | | 主引用文献 | N-terminal structure of ankyrin repeat-containing protein legA11 from Legionella pneumophila
to be published
|
|
4ZXW
 
 | | Crystal structure of SgcC5 protein from Streptomyces globisporus (complex with (R)-(-)-1-(2-naphthyl)-1,2-ethanediol and sucrose) | | 分子名称: | (1R)-1-(naphthalen-2-yl)ethane-1,2-diol, 3-CYCLOHEXYL-1-PROPYLSULFONIC ACID, C-domain type II peptide synthetase, ... | | 著者 | Michalska, K, Bigelow, L, Jedrzejczak, R, Babnigg, G, Lohman, J, Ma, M, Rudolf, J, Chang, C.-Y, Shen, B, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | 登録日 | 2015-05-20 | | 公開日 | 2015-06-17 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.187 Å) | | 主引用文献 | Crystal structure of SgcC5 protein from Streptomyces globisporus
To Be Published
|
|
