4N3Y
 
 | |
4N3Z
 
 | |
4N3X
 
 | |
9J4I
 
 | | Crystal structure of GH9l Inulin fructotransferases (IFTase) in compex with fruetosyl nystose (GF4) | | 分子名称: | DFA-III-forming inulin fructotransferase, beta-D-fructofuranose-(2-1)-beta-D-fructofuranose-(2-1)-[alpha-D-glucopyranose-(1-2)]beta-D-fructofuranose, beta-D-fructofuranose-(2-1)-beta-D-fructofuranose-(2-1)-beta-D-fructofuranose-(2-1)-[alpha-D-glucopyranose-(1-2)]beta-D-fructofuranose | | 著者 | Chen, G, Wang, Z.X, Yang, Y.Q, Li, Y.G, Zhang, T, Ouyang, S.Y, Zhang, L, Chen, Y, Ruan, X.L, Miao, M. | | 登録日 | 2024-08-09 | | 公開日 | 2024-09-04 | | 最終更新日 | 2024-09-18 | | 実験手法 | X-RAY DIFFRACTION (1.96 Å) | | 主引用文献 | Elucidation of the mechanism underlying the sequential catalysis of inulin by fructotransferase.
Int.J.Biol.Macromol., 277, 2024
|
|
9J4K
 
 | | Crystal structure of GH9l Inulinfructotransferases (IFTase) in complex with GF2 | | 分子名称: | DFA-III-forming inulin fructotransferase, beta-D-fructofuranose-(2-1)-beta-D-fructofuranose-(2-1)-alpha-D-glucopyranose | | 著者 | Chen, G, Wang, Z.X, Yang, Y.Q, Li, Y.G, Zhang, T, Ouyang, S.Y, Zhang, L, Chen, Y, Ruan, X.L, Miao, M. | | 登録日 | 2024-08-09 | | 公開日 | 2024-09-04 | | 最終更新日 | 2024-09-18 | | 実験手法 | X-RAY DIFFRACTION (2.201 Å) | | 主引用文献 | Elucidation of the mechanism underlying the sequential catalysis of inulin by fructotransferase.
Int.J.Biol.Macromol., 277, 2024
|
|
9J4J
 
 | | Crystal structure of GH9l Inulin fructotransferases(IFTase)incomplex with nystose(F3) | | 分子名称: | DFA-III-forming inulin fructotransferase, beta-D-fructofuranose, beta-D-fructofuranose-(1-1)-beta-D-fructofuranose, ... | | 著者 | Chen, G, Wang, Z.X, Yang, Y.Q, Li, Y.G, Zhang, T, Ouyang, S.Y, Zhang, L, Chen, Y, Ruan, X.L, Miao, M. | | 登録日 | 2024-08-09 | | 公開日 | 2024-09-04 | | 最終更新日 | 2024-09-18 | | 実験手法 | X-RAY DIFFRACTION (2.803 Å) | | 主引用文献 | Elucidation of the mechanism underlying the sequential catalysis of inulin by fructotransferase.
Int.J.Biol.Macromol., 277, 2024
|
|
9J4L
 
 | | Crystal structure of GH9l Inulin fructotransferases (IFTase) | | 分子名称: | DFA-III-forming inulin fructotransferase | | 著者 | Chen, G, Wang, Z.X, Yang, Y.Q, Li, Y.G, Zhang, T, Ouyang, S.Y, Zhang, L, Chen, Y, Ruan, X.L, Miao, M. | | 登録日 | 2024-08-09 | | 公開日 | 2024-09-04 | | 最終更新日 | 2024-09-18 | | 実験手法 | X-RAY DIFFRACTION (2.15 Å) | | 主引用文献 | Elucidation of the mechanism underlying the sequential catalysis of inulin by fructotransferase.
Int.J.Biol.Macromol., 277, 2024
|
|
1QXK
 
 | | Monoacid-Based, Cell Permeable, Selective Inhibitors of Protein Tyrosine Phosphatase 1B | | 分子名称: | 2-{4-[2-ACETYLAMINO-3-(4-CARBOXYMETHOXY-3-HYDROXY-PHENYL)-PROPIONYLAMINO]-BUTOXY}-6-HYDROXY-BENZOIC ACID METHYL ESTER, Protein-tyrosine phosphatase, non-receptor type 1 | | 著者 | Xin, Z, Liu, G, Abad-Zapatero, C, Pei, Z, Szczepankiewick, B.G, Li, X, Zhang, T, Hutchins, C.W, Hajduk, P.J, Ballaron, S.J, Stashko, M.A, Lubben, T.H, Trevillyan, J.M, Jirousek, M.R. | | 登録日 | 2003-09-08 | | 公開日 | 2003-10-28 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Identification of a Monoacid-Based, Cell Permeable, Selective
Inhibitor of Protein Tyrosine Phosphatase 1B
BIOORG.MED.CHEM.LETT., 13, 2003
|
|
5C77
 
 | | A novel protein arginine methyltransferase | | 分子名称: | Protein arginine N-methyltransferase SFM1, S-ADENOSYL-L-HOMOCYSTEINE | | 著者 | Lv, F, Zhang, T, Ding, J. | | 登録日 | 2015-06-24 | | 公開日 | 2016-01-13 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Structural basis for Sfm1 functioning as a protein arginine methyltransferase.
Cell Discov, 1, 2015
|
|
5C3Q
 
 | | Crystal structure of the full-length Neurospora crassa T7H in complex with alpha-KG and thymine (T) | | 分子名称: | 1,2-ETHANEDIOL, 2-OXOGLUTARIC ACID, NICKEL (II) ION, ... | | 著者 | Li, W, Zhang, T, Ding, J. | | 登録日 | 2015-06-17 | | 公開日 | 2015-10-21 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.05 Å) | | 主引用文献 | Molecular basis for the substrate specificity and catalytic mechanism of thymine-7-hydroxylase in fungi
Nucleic Acids Res., 43, 2015
|
|
5J5C
 
 | | Crystal structure of ARL1-GTP and DCB domain of BIG1 complex | | 分子名称: | ADP-ribosylation factor-like protein 1, Brefeldin A-inhibited guanine nucleotide-exchange protein 1, GUANOSINE-5'-TRIPHOSPHATE, ... | | 著者 | Wang, R, Wang, Z, Zhang, T, Ding, J. | | 登録日 | 2016-04-02 | | 公開日 | 2016-09-28 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (3.4 Å) | | 主引用文献 | Structural basis for targeting BIG1 to Golgi apparatus through interaction of its DCB domain with Arl1
J Mol Cell Biol, 2016
|
|
5C3O
 
 | | Crystal structure of the C-terminal truncated Neurospora crassa T7H (NcT7HdeltaC) in apo form | | 分子名称: | 1,2-ETHANEDIOL, CALCIUM ION, Thymine dioxygenase | | 著者 | Li, W, Zhang, T, Ding, J. | | 登録日 | 2015-06-17 | | 公開日 | 2015-10-21 | | 最終更新日 | 2015-12-02 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Molecular basis for the substrate specificity and catalytic mechanism of thymine-7-hydroxylase in fungi
Nucleic Acids Res., 43, 2015
|
|
5C3R
 
 | | Crystal structure of the full-length Neurospora crassa T7H in complex with alpha-KG and 5-hydroxymethyluracil (5hmU) | | 分子名称: | 1,2-ETHANEDIOL, 2-OXOGLUTARIC ACID, 5-HYDROXYMETHYL URACIL, ... | | 著者 | Li, W, Zhang, T, Ding, J. | | 登録日 | 2015-06-17 | | 公開日 | 2015-10-21 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.35 Å) | | 主引用文献 | Molecular basis for the substrate specificity and catalytic mechanism of thymine-7-hydroxylase in fungi
Nucleic Acids Res., 43, 2015
|
|
5C3S
 
 | | Crystal structure of the full-length Neurospora crassa T7H in complex with alpha-KG and 5-formyluracil (5fU) | | 分子名称: | 1,2-ETHANEDIOL, 2,4-dioxo-1,2,3,4-tetrahydropyrimidine-5-carbaldehyde, 2-OXOGLUTARIC ACID, ... | | 著者 | Li, W, Zhang, T, Ding, J. | | 登録日 | 2015-06-17 | | 公開日 | 2015-10-21 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.15 Å) | | 主引用文献 | Molecular basis for the substrate specificity and catalytic mechanism of thymine-7-hydroxylase in fungi
Nucleic Acids Res., 43, 2015
|
|
5C3P
 
 | | Crystal structure of the full-length Neurospora crassa T7H in complex with alpha-KG | | 分子名称: | 1,2-ETHANEDIOL, 2-OXOGLUTARIC ACID, NICKEL (II) ION, ... | | 著者 | Li, W, Zhang, T, Ding, J. | | 登録日 | 2015-06-17 | | 公開日 | 2015-10-21 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Molecular basis for the substrate specificity and catalytic mechanism of thymine-7-hydroxylase in fungi
Nucleic Acids Res., 43, 2015
|
|
5XYN
 
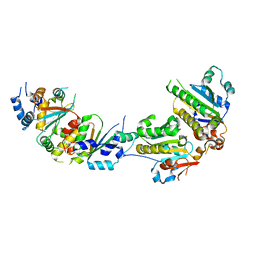 | | The crystal structure of Csm2-Psy3-Shu1-Shu2 complex from budding yeast | | 分子名称: | Chromosome segregation in meiosis protein 2, Platinum sensitivity protein 3, Suppressor of HU sensitivity involved in recombination protein 1, ... | | 著者 | Zhang, S, Zhang, T, Ding, J. | | 登録日 | 2017-07-09 | | 公開日 | 2017-11-08 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (3.3 Å) | | 主引用文献 | Structural basis for the functional role of the Shu complex in homologous recombination.
Nucleic Acids Res., 45, 2017
|
|
4X9Z
 
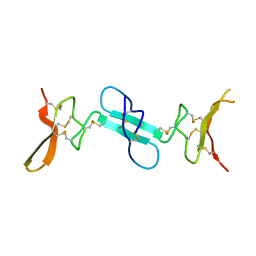 | | Dimeric conotoxin alphaD-GeXXA | | 分子名称: | alphaD-conotoxin GeXXA from the venom of Conus generalis | | 著者 | Xu, S, Zhang, T, Kompella, S, Adams, D, Ding, J, Wang, C. | | 登録日 | 2014-12-12 | | 公開日 | 2015-12-02 | | 最終更新日 | 2020-02-05 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Conotoxin alpha D-GeXXA utilizes a novel strategy to antagonize nicotinic acetylcholine receptors
Sci Rep, 5, 2015
|
|
4XPM
 
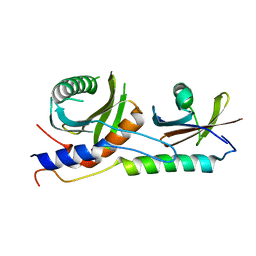 | | Crystal structure of EGO-TC | | 分子名称: | Protein MEH1, Protein SLM4, Uncharacterized protein YCR075W-A | | 著者 | Powis, K, Zhang, T, De Virgilio, C, Ding, J. | | 登録日 | 2015-01-17 | | 公開日 | 2015-08-05 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Crystal structure of the Ego1-Ego2-Ego3 complex and its role in promoting Rag GTPase-dependent TORC1 signaling.
Cell Res., 25, 2015
|
|
3OV6
 
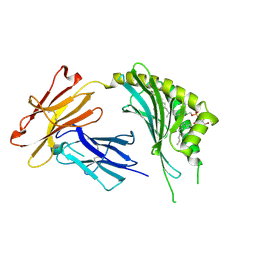 | | CD1c in complex with MPM (mannosyl-beta1-phosphomycoketide) | | 分子名称: | 1-O-[(S)-hydroxy{[(4S,8S,16S,20S)-4,8,12,16,20-pentamethylheptacosyl]oxy}phosphoryl]-beta-D-mannopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-2-microglobulin, ... | | 著者 | Scharf, L, Li, N.S, Hawk, A.J, Garzon, D, Zhang, T, Kazen, A.R, Shah, S, Haddadian, E.J, Saghatelian, A, Faraldo-Gomez, J.D, Meredith, S.C, Piccirilli, J.A, Adams, E.J. | | 登録日 | 2010-09-15 | | 公開日 | 2011-01-19 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (2.502 Å) | | 主引用文献 | The 2.5 A structure of CD1c in complex with a mycobacterial lipid reveals an open groove ideally suited for diverse antigen presentation
Immunity, 33, 2010
|
|
6LQI
 
 | | Cryo-EM structure of the mouse Piezo1 isoform Piezo1.1 | | 分子名称: | Piezo-type mechanosensitive ion channel component 1 | | 著者 | Geng, J, Liu, W, Zhou, H, Zhang, T, Wang, L, Zhang, M, Shen, B, Li, X, Xiao, B. | | 登録日 | 2020-01-13 | | 公開日 | 2020-03-04 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (4.5 Å) | | 主引用文献 | A Plug-and-Latch Mechanism for Gating the Mechanosensitive Piezo Channel.
Neuron, 106, 2020
|
|
6BNB
 
 | | Crystal structure of DDB1-CRBN-BRD4(BD1) complex bound to dBET57 PROTAC | | 分子名称: | Bromodomain-containing protein 4, DNA damage-binding protein 1, Protein cereblon, ... | | 著者 | Nowak, R.P, DeAngelo, S.L, Buckley, D, Ishoey, M, He, Z, Zhang, T, Bradner, J.E, Fischer, E.S. | | 登録日 | 2017-11-16 | | 公開日 | 2018-05-30 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (6.343 Å) | | 主引用文献 | Plasticity in binding confers selectivity in ligand-induced protein degradation.
Nat. Chem. Biol., 14, 2018
|
|
6LRU
 
 | | Marsupenaeus japonicus ferritin mutant (T158H) | | 分子名称: | FE (III) ION, Ferritin, IMIDAZOLE, ... | | 著者 | Zhao, G, Tan, X, Zhang, T. | | 登録日 | 2020-01-16 | | 公開日 | 2020-11-25 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Converting histidine-induced 3D protein arrays in crystals into their 3D analogues in solution by metal coordination cross-linking.
Commun Chem, 2020
|
|
6LRV
 
 | |
6LRW
 
 | |
6LS2
 
 | |
