5GZ4
 
 | |
5GZ5
 
 | |
5HI7
 
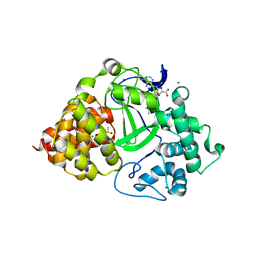 | | Co-crystal structure of human SMYD3 with an aza-SAH compound | | 分子名称: | 5'-{[(3S)-3-amino-3-carboxypropyl][3-(dimethylamino)propyl]amino}-5'-deoxyadenosine, DIMETHYL SULFOXIDE, Histone-lysine N-methyltransferase SMYD3, ... | | 著者 | Elkins, P.A, Bonnette, W.G. | | 登録日 | 2016-01-11 | | 公開日 | 2016-03-30 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (2.15 Å) | | 主引用文献 | Structure-Based Design of a Novel SMYD3 Inhibitor that Bridges the SAM-and MEKK2-Binding Pockets.
Structure, 24, 2016
|
|
5I7J
 
 | |
5I7K
 
 | | Crystal Structure of Human SPLUNC1 Dolphin Mutant D1 (G58A, S61A, G62E, G63D, G66D, I67T) | | 分子名称: | BPI fold-containing family A member 1 | | 著者 | Walton, W.G, Redinbo, M.R. | | 登録日 | 2016-02-17 | | 公開日 | 2016-05-18 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.552 Å) | | 主引用文献 | Structural Features Essential to the Antimicrobial Functions of Human SPLUNC1.
Biochemistry, 55, 2016
|
|
5HQ8
 
 | |
5I7L
 
 | |
4RE9
 
 | | Crystal structure of human insulin degrading enzyme (IDE) in complex with compound 71290 | | 分子名称: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, 4-fluoro-N-({1-[(2R)-4-(hydroxyamino)-1-(naphthalen-2-yl)-4-oxobutan-2-yl]-1H-1,2,3-triazol-5-yl}methyl)benzamide, ... | | 著者 | Liang, W.G, Deprez, R, Deprez, B, Tang, W.J. | | 登録日 | 2014-09-22 | | 公開日 | 2015-09-30 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.908 Å) | | 主引用文献 | Catalytic site inhibition of insulin-degrading enzyme by a small molecule induces glucose intolerance in mice.
Nat Commun, 6, 2015
|
|
4RA8
 
 | | Structure analysis of the Mip1a P8A mutant | | 分子名称: | C-C motif chemokine 3 | | 著者 | Liang, W.G, Ren, M, Guo, Q, Tang, W.J. | | 登録日 | 2014-09-09 | | 公開日 | 2014-09-24 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Structures of human CCL18, CCL3, and CCL4 reveal molecular determinants for quaternary structures and sensitivity to insulin-degrading enzyme.
J.Mol.Biol., 427, 2015
|
|
4RAL
 
 | | Crystal structure of insulin degrading enzyme in complex with macrophage inflammatory protein 1 beta | | 分子名称: | C-C motif chemokine 4, Insulin-degrading enzyme, ZINC ION | | 著者 | Liang, W.G, Ren, M, Guo, Q, Tang, W.J. | | 登録日 | 2014-09-10 | | 公開日 | 2015-05-13 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (3.148 Å) | | 主引用文献 | Structures of human CCL18, CCL3, and CCL4 reveal molecular determinants for quaternary structures and sensitivity to insulin-degrading enzyme.
J.Mol.Biol., 427, 2015
|
|
4RJ1
 
 | | Structural variations and solvent structure of UGGGGU quadruplexes stabilized by Sr2+ ions | | 分子名称: | CALCIUM ION, RNA (5'-R(*UP*GP*GP*GP*GP*U)-3'), SODIUM ION, ... | | 著者 | Fyfe, A.C, Dunten, P.W, Scott, W.G. | | 登録日 | 2014-10-08 | | 公開日 | 2014-11-12 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (0.92 Å) | | 主引用文献 | Structural Variations and Solvent Structure of r(UGGGGU) Quadruplexes Stabilized by Sr(2+) Ions.
J.Mol.Biol., 427, 2015
|
|
4RKV
 
 | | Structural variations and solvent structure of UGGGGU quadruplexes stabilized by Sr2+ ions | | 分子名称: | CALCIUM ION, RNA (5'-R(*UP*GP*GP*GP*GP*U)-3'), SODIUM ION, ... | | 著者 | Fyfe, A.C, Dunten, P.W, Scott, W.G. | | 登録日 | 2014-10-14 | | 公開日 | 2014-11-19 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (0.88 Å) | | 主引用文献 | Structural Variations and Solvent Structure of r(UGGGGU) Quadruplexes Stabilized by Sr(2+) Ions.
J.Mol.Biol., 427, 2015
|
|
4RPU
 
 | | Crystal Structure of Human Presequence Protease in Complex with Inhibitor MitoBloCK-60 | | 分子名称: | ACETATE ION, CALCIUM ION, GLYCEROL, ... | | 著者 | Mo, S.M, Liang, W.G, King, J.V, Wijaya, J, Koehler, C.M, Tang, W.J. | | 登録日 | 2014-10-31 | | 公開日 | 2015-12-09 | | 実験手法 | X-RAY DIFFRACTION (2.265 Å) | | 主引用文献 | Crystal Structure of Human Presequence Protease in Complex with Inhibitor MitoBloCK-60
TO BE PUBLISHED
|
|
4RNE
 
 | | Structural variations and solvent structure of UGGGGU quadruplexes stabilized by Sr2+ ions | | 分子名称: | CALCIUM ION, RNA (5'-R(*UP*GP*GP*GP*GP*U)-3'), SODIUM ION, ... | | 著者 | Fyfe, A.C, Dunten, P.W, Scott, W.G. | | 登録日 | 2014-10-24 | | 公開日 | 2014-11-19 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.01 Å) | | 主引用文献 | Structural Variations and Solvent Structure of r(UGGGGU) Quadruplexes Stabilized by Sr(2+) Ions.
J.Mol.Biol., 427, 2015
|
|
1NNB
 
 | | THREE-DIMENSIONAL STRUCTURE OF INFLUENZA A N9 NEURAMINIDASE AND ITS COMPLEX WITH THE INHIBITOR 2-DEOXY 2,3-DEHYDRO-N-ACETYL NEURAMINIC ACID | | 分子名称: | 2-DEOXY-2,3-DEHYDRO-N-ACETYL-NEURAMINIC ACID, CALCIUM ION, NEURAMINIDASE | | 著者 | Bossart-Whitaker, P, Carson, M, Babu, Y.S, Smith, C.D, Laver, W.G, Air, G.M. | | 登録日 | 1993-03-08 | | 公開日 | 1994-04-30 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Three-dimensional structure of influenza A N9 neuraminidase and its complex with the inhibitor 2-deoxy 2,3-dehydro-N-acetyl neuraminic acid.
J.Mol.Biol., 232, 1993
|
|
1YF5
 
 | | Cyto-Epsl: The Cytoplasmic Domain Of Epsl, An Inner Membrane Component Of The Type II Secretion System Of Vibrio Cholerae | | 分子名称: | General secretion pathway protein L | | 著者 | Abendroth, J, Murphy, P, Mushtaq, A, Sandkvist, M, Bagdasarian, M, Hol, W.G. | | 登録日 | 2004-12-30 | | 公開日 | 2005-05-03 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.75 Å) | | 主引用文献 | The X-ray Structure of the Type II Secretion System Complex Formed by the N-terminal Domain of EpsE and the Cytoplasmic Domain of EpsL of Vibrio cholerae.
J.Mol.Biol., 348, 2005
|
|
1NNA
 
 | | THREE-DIMENSIONAL STRUCTURE OF INFLUENZA A N9 NEURAMINIDASE AND ITS COMPLEX WITH THE INHIBITOR 2-DEOXY 2,3-DEHYDRO-N-ACETYL NEURAMINIC ACID | | 分子名称: | CALCIUM ION, NEURAMINIDASE | | 著者 | Bossart-Whitaker, P, Carson, M, Babu, Y.S, Smith, C.D, Laver, W.G, Air, G.M. | | 登録日 | 1993-03-08 | | 公開日 | 1994-04-30 | | 最終更新日 | 2017-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Three-dimensional structure of influenza A N9 neuraminidase and its complex with the inhibitor 2-deoxy 2,3-dehydro-N-acetyl neuraminic acid.
J.Mol.Biol., 232, 1993
|
|
4NN9
 
 | | REFINED ATOMIC STRUCTURES OF N9 SUBTYPE INFLUENZA VIRUS NEURAMINIDASE AND ESCAPE MUTANTS | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, NEURAMINIDASE N9, ... | | 著者 | Tulip, W.R, Varghese, J.N, Baker, A.T, Vandonkelaar, A, Laver, W.G, Webster, R.G, Colman, P.M. | | 登録日 | 1991-03-28 | | 公開日 | 1992-07-15 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Refined atomic structures of N9 subtype influenza virus neuraminidase and escape mutants.
J.Mol.Biol., 221, 1991
|
|
1BI0
 
 | | STRUCTURE OF APO-AND HOLO-DIPHTHERIA TOXIN REPRESSOR | | 分子名称: | DIPHTHERIA TOXIN REPRESSOR, SULFATE ION, ZINC ION | | 著者 | Pohl, E, Hol, W.G. | | 登録日 | 1998-06-21 | | 公開日 | 1999-07-22 | | 最終更新日 | 2022-12-21 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Motion of the DNA-binding domain with respect to the core of the diphtheria toxin repressor (DtxR) revealed in the crystal structures of apo- and holo-DtxR.
J.Biol.Chem., 273, 1998
|
|
2A5F
 
 | | Cholera toxin A1 subunit bound to its substrate, NAD+, and its human protein activator, ARF6 | | 分子名称: | ADP-ribosylation factor 6, Cholera enterotoxin, A chain, ... | | 著者 | O'Neal, C.J, Jobling, M.G, Holmes, R.K, Hol, W.G.J. | | 登録日 | 2005-06-30 | | 公開日 | 2005-08-16 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.02 Å) | | 主引用文献 | Structural basis for the activation of cholera toxin by human ARF6-GTP.
Science, 309, 2005
|
|
301D
 
 | | CAPTURING THE STRUCTURE OF A CATALYTIC RNA INTERMEDIATE: RNA HAMMERHEAD RIBOZYME, MG(II)-SOAKED | | 分子名称: | MAGNESIUM ION, RNA HAMMERHEAD RIBOZYME | | 著者 | Scott, W.G, Murray, J.B, Arnold, J.R.P, Stoddard, B.L, Klug, A. | | 登録日 | 1996-12-14 | | 公開日 | 1997-01-24 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Capturing the structure of a catalytic RNA intermediate: the hammerhead ribozyme.
Science, 274, 1996
|
|
379D
 
 | | THE STRUCTURAL BASIS OF HAMMERHEAD RIBOZYME SELF-CLEAVAGE | | 分子名称: | COBALT (II) ION, RNA (5'-R(*GP*GP*CP*CP*GP*AP*AP*AP*CP*UP*CP*GP*UP*AP*AP*GP*A P*GP*UP*CP*AP*CP*CP*AP*C)-3'), RNA (5'-R(*GP*UP*GP*GP*UP*CP*UP*GP*AP*UP*GP*AP*GP*GP*CP*C)-3') | | 著者 | Murray, J.B, Terwey, D.P, Maloney, L, Karpeisky, A, Usman, N, Beigelman, L, Scott, W.G. | | 登録日 | 1998-02-05 | | 公開日 | 1998-02-11 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (3.1 Å) | | 主引用文献 | The structural basis of hammerhead ribozyme self-cleavage.
Cell(Cambridge,Mass.), 92, 1998
|
|
300D
 
 | | CAPTURING THE STRUCTURE OF A CATALYTIC RNA INTERMEDIATE: RNA HAMMERHEAD RIBOZYME, MN(II)-SOAKED | | 分子名称: | MANGANESE (II) ION, RNA HAMMERHEAD RIBOZYME | | 著者 | Scott, W.G, Murray, J.B, Arnold, J.R.P, Stoddard, B.L, Klug, A. | | 登録日 | 1996-12-14 | | 公開日 | 1997-01-24 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Capturing the structure of a catalytic RNA intermediate: the hammerhead ribozyme.
Science, 274, 1996
|
|
2PHH
 
 | |
6TVI
 
 | | Salmonella typhimurium mutant neuraminidase (D100S)+ DANA | | 分子名称: | 2-DEOXY-2,3-DEHYDRO-N-ACETYL-NEURAMINIC ACID, GLYCEROL, Sialidase | | 著者 | Garman, E.F, Salinger, M.T, Murray, J.W, Laver, W.G, Kuhn, P, Vimr, E.R. | | 登録日 | 2020-01-09 | | 公開日 | 2020-02-12 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1 Å) | | 主引用文献 | Salmonella typhimurium mutant neuraminidase (D100S)+ DANA
To Be Published
|
|
