4IBB
 
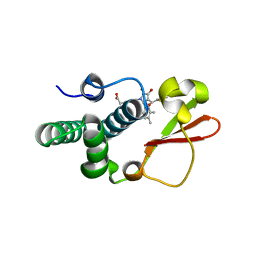 | | Ebola virus VP35 bound to small molecule | | 分子名称: | Polymerase cofactor VP35, {4-[(5R)-3-hydroxy-2-oxo-4-(thiophen-2-ylcarbonyl)-5-(2,4,5-trimethylphenyl)-2,5-dihydro-1H-pyrrol-1-yl]phenyl}acetic acid | | 著者 | Brown, C.S, Leung, D.W, Xu, W, Borek, D.M, Otwinowski, Z, Ramanan, P, Stubbs, A.J, Peterson, D.S, Binning, J.M, Amarasinghe, G.K, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2012-12-08 | | 公開日 | 2014-02-26 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.752 Å) | | 主引用文献 | In Silico Derived Small Molecules Bind the Filovirus VP35 Protein and Inhibit Its Polymerase Cofactor Activity.
J.Mol.Biol., 426, 2014
|
|
4IBD
 
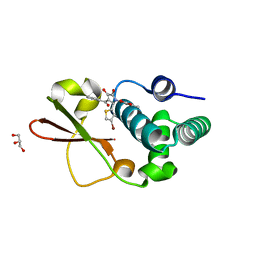 | | Ebola virus VP35 bound to small molecule | | 分子名称: | 5-[(2R)-3-benzoyl-2-(4-bromothiophen-2-yl)-4-hydroxy-5-oxo-2,5-dihydro-1H-pyrrol-1-yl]-2-methylbenzoic acid, GLYCEROL, MAGNESIUM ION, ... | | 著者 | Brown, C.S, Leung, D.W, Xu, W, Borek, D.M, Otwinowski, Z, Ramanan, P, Stubbs, A.J, Peterson, D.S, Binning, J.M, Amarasinghe, G.K. | | 登録日 | 2012-12-08 | | 公開日 | 2014-03-19 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.84 Å) | | 主引用文献 | In Silico Derived Small Molecules Bind the Filovirus VP35 Protein and Inhibit Its Polymerase Cofactor Activity.
J.Mol.Biol., 426, 2014
|
|
4IBJ
 
 | | Ebola virus VP35 bound to small molecule | | 分子名称: | 3-{(5S)-3-hydroxy-2-oxo-4-[3-(trifluoromethyl)benzoyl]-5-[3-(trifluoromethyl)phenyl]-2,5-dihydro-1H-pyrrol-1-yl}benzoic acid, Polymerase cofactor VP35 | | 著者 | Brown, C.S, Leung, D.W, Xu, W, Borek, D.M, Otwinowski, Z, Ramanan, P, Stubbs, A.J, Peterson, D.S, Binning, J.M, Amarasinghe, G.K. | | 登録日 | 2012-12-08 | | 公開日 | 2014-03-19 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.54 Å) | | 主引用文献 | In Silico Derived Small Molecules Bind the Filovirus VP35 Protein and Inhibit Its Polymerase Cofactor Activity.
J.Mol.Biol., 426, 2014
|
|
4IBI
 
 | | Ebola virus VP35 bound to small molecule | | 分子名称: | 3-{(2S)-2-(7-chloro-1,3-benzodioxol-5-yl)-4-hydroxy-5-oxo-3-[3-(trifluoromethyl)benzoyl]-2,5-dihydro-1H-pyrrol-1-yl}benzoic acid, Polymerase cofactor VP35 | | 著者 | Brown, C.S, Leung, D.W, Xu, W, Borek, D.M, Otwinowski, Z, Ramanan, P, Stubbs, A.J, Peterson, D.S, Binning, J.M, Amarasinghe, G.K. | | 登録日 | 2012-12-08 | | 公開日 | 2014-03-19 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.473 Å) | | 主引用文献 | In Silico Derived Small Molecules Bind the Filovirus VP35 Protein and Inhibit Its Polymerase Cofactor Activity.
J.Mol.Biol., 426, 2014
|
|
4IBC
 
 | | Ebola virus VP35 bound to small molecule | | 分子名称: | DIMETHYL SULFOXIDE, Polymerase cofactor VP35, {4-[(2R)-3-(2-chlorobenzoyl)-2-(2-chlorophenyl)-4-hydroxy-5-oxo-2,5-dihydro-1H-pyrrol-1-yl]phenyl}acetic acid | | 著者 | Brown, C.S, Leung, D.W, Xu, W, Borek, D.M, Otwinowski, Z, Ramanan, P, Stubbs, A.J, Peterson, D.S, Binning, J.M, Amarasinghe, G.K. | | 登録日 | 2012-12-08 | | 公開日 | 2014-03-19 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.745 Å) | | 主引用文献 | In Silico Derived Small Molecules Bind the Filovirus VP35 Protein and Inhibit Its Polymerase Cofactor Activity.
J.Mol.Biol., 426, 2014
|
|
7KQ8
 
 | | Structure of iron bound MEMO1 | | 分子名称: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, FE (II) ION, ... | | 著者 | Boniecki, M.T, Uhlemann, E.E, Dmitriev, O.Y. | | 登録日 | 2020-11-13 | | 公開日 | 2021-11-17 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (2.15 Å) | | 主引用文献 | MEMO1 binds iron and modulates iron homeostasis in cancer cells.
Elife, 13, 2024
|
|
7L5C
 
 | | Structure of copper bound MEMO1 | | 分子名称: | 1,2-ETHANEDIOL, COPPER (I) ION, GLYCEROL, ... | | 著者 | Boniecki, M.T, Uhlemann, E.E, Dmitriev, O.Y. | | 登録日 | 2020-12-21 | | 公開日 | 2022-01-19 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (2.55 Å) | | 主引用文献 | MEMO1 binds iron and modulates iron homeostasis in cancer cells.
Elife, 13, 2024
|
|
3FSM
 
 | |
4IBE
 
 | | Ebola virus VP35 bound to small molecule | | 分子名称: | 5-[(2R)-3-benzoyl-2-(4-bromothiophen-2-yl)-4-hydroxy-5-oxo-2,5-dihydro-1H-pyrrol-1-yl]-2-chlorobenzoic acid, GLYCEROL, Polymerase cofactor VP35 | | 著者 | Brown, C.S, Leung, D.W, Xu, W, Borek, D.M, Otwinowski, Z, Ramanan, P, Stubbs, A.J, Peterson, D.S, Binning, J.M, Amarasinghe, G.K, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2012-12-08 | | 公開日 | 2014-03-19 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | In Silico Derived Small Molecules Bind the Filovirus VP35 Protein and Inhibit Its Polymerase Cofactor Activity.
J.Mol.Biol., 426, 2014
|
|
4IBF
 
 | | Ebola virus VP35 bound to small molecule | | 分子名称: | (4-{(2R)-2-(4-bromothiophen-2-yl)-3-[(5-chlorothiophen-2-yl)carbonyl]-4-hydroxy-5-oxo-2,5-dihydro-1H-pyrrol-1-yl}phenyl)acetic acid, Polymerase cofactor VP35 | | 著者 | Brown, C.S, Leung, D.W, Xu, W, Borek, D.M, Otwinowski, Z, Ramanan, P, Stubbs, A.J, Peterson, D.S, Binning, J.M, Amarasinghe, G.K, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2012-12-08 | | 公開日 | 2014-03-19 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.291 Å) | | 主引用文献 | In Silico Derived Small Molecules Bind the Filovirus VP35 Protein and Inhibit Its Polymerase Cofactor Activity.
J.Mol.Biol., 426, 2014
|
|
4IBK
 
 | | Ebola virus VP35 bound to small molecule | | 分子名称: | 3-{(2S)-2-(7-chloro-1,3-benzodioxol-5-yl)-3-[(5-chlorothiophen-2-yl)carbonyl]-4-hydroxy-5-oxo-2,5-dihydro-1H-pyrrol-1-yl}benzoic acid, DIMETHYL SULFOXIDE, GLYCEROL, ... | | 著者 | Brown, C.S, Leung, D.W, Xu, W, Borek, D.M, Otwinowski, Z, Ramanan, P, Stubbs, A.J, Peterson, D.S, Binning, J.M, Amarasinghe, G.K. | | 登録日 | 2012-12-08 | | 公開日 | 2014-03-19 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | In Silico Derived Small Molecules Bind the Filovirus VP35 Protein and Inhibit Its Polymerase Cofactor Activity.
J.Mol.Biol., 426, 2014
|
|
7M8H
 
 | | Structure of Memo1 C244S metal binding site mutant at 1.75A | | 分子名称: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, DI(HYDROXYETHYL)ETHER, ... | | 著者 | Boniecki, M.T, Uhlemann, E.E, Dmitriev, O.Y. | | 登録日 | 2021-03-29 | | 公開日 | 2022-04-06 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | MEMO1 binds iron and modulates iron homeostasis in cancer cells.
Elife, 13, 2024
|
|
7MKT
 
 | | Crystal structure of r(GU)11G-NMM complex | | 分子名称: | N-METHYLMESOPORPHYRIN, POTASSIUM ION, RNA (5'-R(*GP*UP*GP*UP*GP*UP*GP*UP*GP*UP*GP*UP*GP*UP*GP*UP*GP*UP*GP*UP*GP*UP*G)-3') | | 著者 | Bingman, C.A, Roschdi, S, Nomura, Y, Butcher, S.E. | | 登録日 | 2021-04-26 | | 公開日 | 2022-11-02 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (1.97 Å) | | 主引用文献 | An atypical RNA quadruplex marks RNAs as vectors for gene silencing.
Nat.Struct.Mol.Biol., 29, 2022
|
|
3HAU
 
 | |
3HBO
 
 | |
3HAW
 
 | |
3HDK
 
 | |
3NWQ
 
 | |
3NXE
 
 | |
3NWX
 
 | | X-ray structure of ester chemical analogue [O-Ile50,O-Ile50']HIV-1 protease complexed with KVS-1 inhibitor | | 分子名称: | N~2~-[(2R,5S)-5-({(2S,3S)-2-[(N-acetyl-L-threonyl)amino]-3-methylpent-4-enoyl}amino)-2-butyl-4,4-dihydroxynonanoyl]-L-glutaminyl-L-argininamide, SULFATE ION, protease | | 著者 | Torbeev, V.Y, Kent, S.B.H. | | 登録日 | 2010-07-12 | | 公開日 | 2011-11-02 | | 最終更新日 | 2019-07-31 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Protein conformational dynamics in the mechanism of HIV-1 protease catalysis.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3NXN
 
 | | X-ray structure of ester chemical analogue 'covalent dimer' [Ile50,O-Ile50']HIV-1 protease complexed with KVS-1 inhibitor | | 分子名称: | N~2~-[(2R,5S)-5-({(2S,3S)-2-[(N-acetyl-L-threonyl)amino]-3-methylpent-4-enoyl}amino)-2-butyl-4,4-dihydroxynonanoyl]-L-glutaminyl-L-argininamide, SULFATE ION, protease covalent dimer | | 著者 | Torbeev, V.Y, Kent, S.B.H. | | 登録日 | 2010-07-14 | | 公開日 | 2011-11-02 | | 最終更新日 | 2019-07-31 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Protein conformational dynamics in the mechanism of HIV-1 protease catalysis.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3NYG
 
 | |
3O7L
 
 | |
2EVN
 
 | | NMR solution structures of At1g77540 | | 分子名称: | Protein At1g77540 | | 著者 | Tyler, R.C, Singh, S, Tonelli, M, Min, M.S, Markley, J.L, Center for Eukaryotic Structural Genomics (CESG) | | 登録日 | 2005-10-31 | | 公開日 | 2005-11-15 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structure of Arabidopsis thaliana At1g77540 Protein, a Minimal Acetyltransferase from the COG2388 Family.
Biochemistry, 45, 2006
|
|
8TNS
 
 | |
