4BJ1
 
 | | Crystal structure of Saccharomyces cerevisiae RIF2 | | 分子名称: | CHLORIDE ION, PROTEIN RIF2 | | 著者 | Shi, T, Bunker, R.D, Gut, H, Scrima, A, Thoma, N.H. | | 登録日 | 2013-04-15 | | 公開日 | 2013-06-19 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.94 Å) | | 主引用文献 | Rif1 and Rif2 Shape Telomere Funcation and Architecture Through Multivalent RAP1 Interactions
Cell(Cambridge,Mass.), 153, 2013
|
|
4BJT
 
 | | Crystal structure of the Rap1 C-terminal domain (Rap1-RCT) in complex with the Rap1 binding module of Rif1 (Rif1-RBM) | | 分子名称: | 1,2-ETHANEDIOL, DNA-BINDING PROTEIN RAP1, TELOMERE LENGTH REGULATOR PROTEIN RIF1 | | 著者 | Shi, T, Bunker, R.D, Gut, H, Scrima, A, Thoma, N.H. | | 登録日 | 2013-04-19 | | 公開日 | 2013-06-19 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.61 Å) | | 主引用文献 | Rif1 and Rif2 Shape Telomere Funcation and Architecture Through Multivalent RAP1 Interactions
Cell(Cambridge,Mass.), 153, 2013
|
|
4BJ6
 
 | | Crystal structure Rif2 in complex with the C-terminal domain of Rap1 (Rap1-RCT) | | 分子名称: | DNA-BINDING PROTEIN RAP1, RAP1-INTERACTING FACTOR 2, SULFATE ION | | 著者 | Shi, T, Bunker, R.D, Gut, H, Scrima, A, Thoma, N.H. | | 登録日 | 2013-04-16 | | 公開日 | 2013-06-19 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (3.26 Å) | | 主引用文献 | Rif1 and Rif2 Shape Telomere Funcation and Architecture Through Multivalent RAP1 Interactions
Cell(Cambridge,Mass.), 153, 2013
|
|
4BJS
 
 | | Crystal structure of the Rif1 C-terminal domain (Rif1-CTD) from Saccharomyces cerevisiae | | 分子名称: | TELOMERE LENGTH REGULATOR PROTEIN RIF1 | | 著者 | Bunker, R.D, Shi, T, Gut, H, Scrima, A, Thoma, N.H. | | 登録日 | 2013-04-19 | | 公開日 | 2013-06-19 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (1.94 Å) | | 主引用文献 | Rif1 and Rif2 Shape Telomere Funcation and Architecture Through Multivalent RAP1 Interactions
Cell(Cambridge,Mass.), 153, 2013
|
|
1MIU
 
 | | Structure of a BRCA2-DSS1 complex | | 分子名称: | Breast Cancer type 2 susceptibility protein, Deleted in split hand/split foot protein 1, MERCURY (II) ION | | 著者 | Yang, H, Jeffrey, P.D, Miller, J, Kinnucan, E, Sun, Y, Thoma, N.H, Zheng, N, Chen, P.L, Lee, W.H, Pavletich, N.P. | | 登録日 | 2002-08-23 | | 公開日 | 2002-09-25 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (3.1 Å) | | 主引用文献 | BRCA2 function in DNA binding and recombination from a BRCA2-DSS1-ssDNA
structure
Science, 297, 2002
|
|
3EI2
 
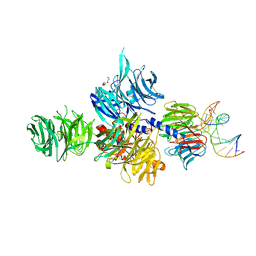 | |
3EI1
 
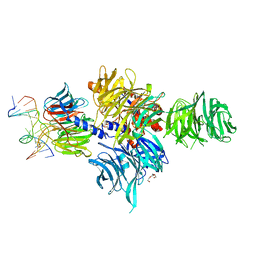 | |
3EI4
 
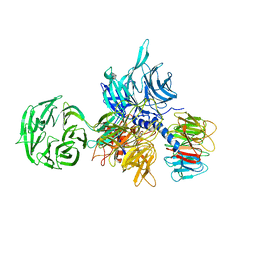 | |
3EI3
 
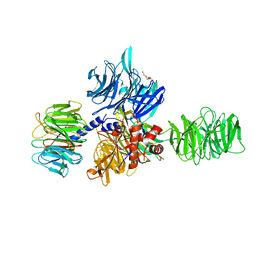 | | Structure of the hsDDB1-drDDB2 complex | | 分子名称: | DNA damage-binding protein 1, DNA damage-binding protein 2, TETRAETHYLENE GLYCOL | | 著者 | Scrima, A, Thoma, N.H. | | 登録日 | 2008-09-15 | | 公開日 | 2009-01-20 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Structural basis of UV DNA-damage recognition by the DDB1-DDB2 complex.
Cell(Cambridge,Mass.), 135, 2008
|
|
2YBA
 
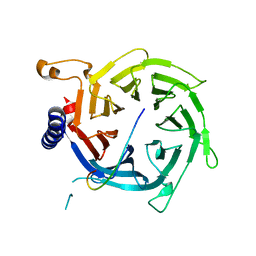 | | Crystal structure of Nurf55 in complex with histone H3 | | 分子名称: | HISTONE H3, PROBABLE HISTONE-BINDING PROTEIN CAF1 | | 著者 | Schmitges, F.W, Prusty, A.B, Faty, M, Stutzer, A, Lingaraju, G.M, Aiwazian, J, Sack, R, Hess, D, Li, L, Zhou, S, Bunker, R.D, Wirth, U, Bouwmeester, T, Bauer, A, Ly-Hartig, N, Zhao, K, Chan, H, Gu, J, Gut, H, Fischle, W, Muller, J, Thoma, N.H. | | 登録日 | 2011-03-02 | | 公開日 | 2011-05-11 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (2.55 Å) | | 主引用文献 | Histone Methylation by Prc2 is Inhibited by Active Chromatin Marks
Mol.Cell, 42, 2011
|
|
4CI2
 
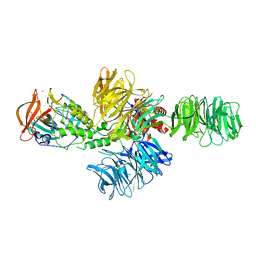 | | Structure of the DDB1-CRBN E3 ubiquitin ligase bound to lenalidomide | | 分子名称: | DNA DAMAGE-BINDING PROTEIN 1, PROTEIN CEREBLON, S-Lenalidomide, ... | | 著者 | Fischer, E.S, Boehm, K, Thoma, N.H. | | 登録日 | 2013-12-05 | | 公開日 | 2014-07-16 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (2.95 Å) | | 主引用文献 | Structure of the Ddb1-Crbn E3 Ubiquitin Ligase in Complex with Thalidomide.
Nature, 512, 2014
|
|
4D18
 
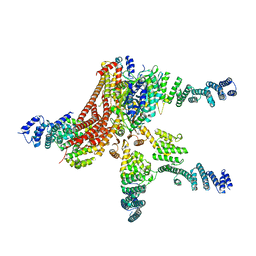 | | Crystal structure of the COP9 signalosome | | 分子名称: | COP9 SIGNALOSOME COMPLEX SUBUNIT 1, COP9 SIGNALOSOME COMPLEX SUBUNIT 2, COP9 SIGNALOSOME COMPLEX SUBUNIT 3, ... | | 著者 | Bunker, R.D, Lingaraju, G.M, Thoma, N.H. | | 登録日 | 2014-05-01 | | 公開日 | 2014-07-23 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (4.08 Å) | | 主引用文献 | Crystal Structure of the Human Cop9 Signalosome
Nature, 512, 2014
|
|
4D0P
 
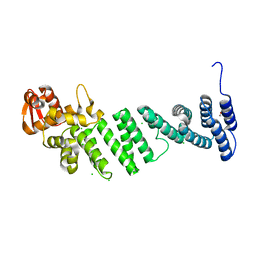 | | Crystal structure of human CSN4 | | 分子名称: | 1,2-ETHANEDIOL, CHLORIDE ION, COP9 SIGNALOSOME COMPLEX SUBUNIT 4, ... | | 著者 | Bunker, R.D, Lingaraju, G.M, Thoma, N.H. | | 登録日 | 2014-04-29 | | 公開日 | 2014-07-23 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Crystal Structure of the Cop9 Signalosome
Nature, 512, 2014
|
|
4D10
 
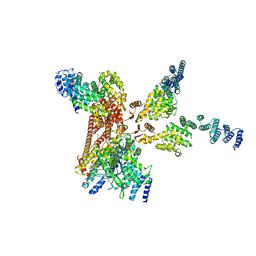 | | Crystal structure of the COP9 signalosome | | 分子名称: | COP9 SIGNALOSOME COMPLEX SUBUNIT 1, COP9 SIGNALOSOME COMPLEX SUBUNIT 2, COP9 SIGNALOSOME COMPLEX SUBUNIT 3, ... | | 著者 | Bunker, R.D, Lingaraju, G.M, Thoma, N.H. | | 登録日 | 2014-04-30 | | 公開日 | 2014-07-23 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (3.8 Å) | | 主引用文献 | Crystal Structure of the Human Cop9 Signalosome
Nature, 512, 2014
|
|
4CI1
 
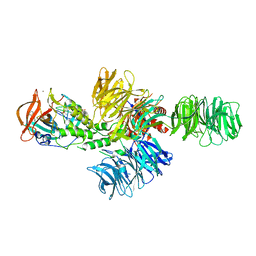 | | Structure of the DDB1-CRBN E3 ubiquitin ligase bound to thalidomide | | 分子名称: | DNA DAMAGE-BINDING PROTEIN 1, PROTEIN CEREBLON, S-Thalidomide, ... | | 著者 | Fischer, E.S, Boehm, K, Thoma, N.H. | | 登録日 | 2013-12-05 | | 公開日 | 2014-07-16 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (2.98 Å) | | 主引用文献 | Structure of the Ddb1-Crbn E3 Ubiquitin Ligase in Complex with Thalidomide.
Nature, 512, 2014
|
|
4CGY
 
 | |
4CI3
 
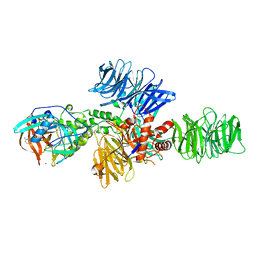 | | Structure of the DDB1-CRBN E3 ubiquitin ligase bound to Pomalidomide | | 分子名称: | DNA DAMAGE-BINDING PROTEIN 1, PROTEIN CEREBLON, S-Pomalidomide, ... | | 著者 | Fischer, E.S, Boehm, K, Thoma, N.H. | | 登録日 | 2013-12-05 | | 公開日 | 2014-07-16 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (3.5 Å) | | 主引用文献 | Structure of the Ddb1-Crbn E3 Ubiquitin Ligase in Complex with Thalidomide.
Nature, 512, 2014
|
|
4CHT
 
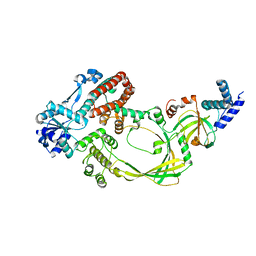 | |
8AJM
 
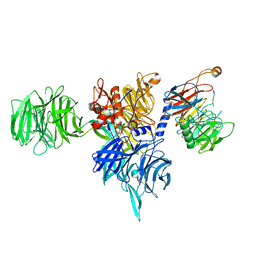 | | Structure of human DDB1-DCAF12 in complex with the C-terminus of CCT5 | | 分子名称: | DDB1- and CUL4-associated factor 12, DNA damage-binding protein 1, T-complex protein 1 subunit epsilon | | 著者 | Pla-Prats, C, Cavadini, S, Kempf, G, Thoma, N.H. | | 登録日 | 2022-07-28 | | 公開日 | 2022-11-09 | | 最終更新日 | 2023-05-24 | | 実験手法 | ELECTRON MICROSCOPY (2.83 Å) | | 主引用文献 | Recognition of the CCT5 di-Glu degron by CRL4 DCAF12 is dependent on TRiC assembly.
Embo J., 42, 2023
|
|
8AJN
 
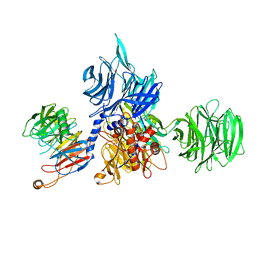 | | Structure of the human DDB1-DCAF12 complex | | 分子名称: | DDB1- and CUL4-associated factor 12, DNA damage-binding protein 1 | | 著者 | Pla-Prats, C, Cavadini, S, Kempf, G, Thoma, N.H. | | 登録日 | 2022-07-28 | | 公開日 | 2022-11-09 | | 最終更新日 | 2023-05-24 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | Recognition of the CCT5 di-Glu degron by CRL4 DCAF12 is dependent on TRiC assembly.
Embo J., 42, 2023
|
|
8AJO
 
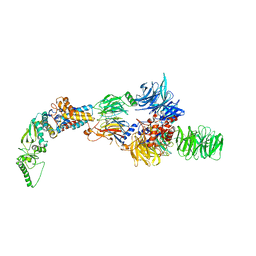 | | Negative-stain electron microscopy structure of DDB1-DCAF12-CCT5 | | 分子名称: | DDB1- and CUL4-associated factor 12, DNA damage-binding protein 1, T-complex protein 1 subunit epsilon | | 著者 | Pla-Prats, C, Cavadini, S, Kempf, G, Thoma, N.H. | | 登録日 | 2022-07-28 | | 公開日 | 2022-11-09 | | 最終更新日 | 2023-05-24 | | 実験手法 | ELECTRON MICROSCOPY (30.6 Å) | | 主引用文献 | Recognition of the CCT5 di-Glu degron by CRL4 DCAF12 is dependent on TRiC assembly.
Embo J., 42, 2023
|
|
8BUM
 
 | | Structure of DDB1 bound to DS15-engaged CDK12-cyclin K | | 分子名称: | (2R)-2-[[6-(5-naphthalen-1-ylpentylamino)-9-propan-2-yl-purin-2-yl]amino]butan-1-ol, Cyclin-K, Cyclin-dependent kinase 12, ... | | 著者 | Kozicka, Z, Kempf, G, Petzold, G, Thoma, N.H. | | 登録日 | 2022-11-30 | | 公開日 | 2023-09-20 | | 最終更新日 | 2024-01-03 | | 実験手法 | X-RAY DIFFRACTION (3.36 Å) | | 主引用文献 | Design principles for cyclin K molecular glue degraders.
Nat.Chem.Biol., 20, 2024
|
|
8BU7
 
 | | Structure of DDB1 bound to 21195-engaged CDK12-cyclin K | | 分子名称: | 1,2-ETHANEDIOL, 1-[2,6-bis(chloranyl)phenyl]-6-[[4-(2-hydroxyethyloxy)phenyl]methyl]-3-propan-2-yl-5H-pyrazolo[3,4-d]pyrimidin-4-one, Cyclin-K, ... | | 著者 | Kozicka, Z, Kempf, G, Petzold, G, Thoma, N.H. | | 登録日 | 2022-11-30 | | 公開日 | 2023-09-13 | | 最終更新日 | 2024-01-03 | | 実験手法 | X-RAY DIFFRACTION (3.245 Å) | | 主引用文献 | Design principles for cyclin K molecular glue degraders.
Nat.Chem.Biol., 20, 2024
|
|
8BUN
 
 | | Structure of DDB1 bound to DS16-engaged CDK12-cyclin K | | 分子名称: | (2~{R})-2-[[6-[(4-phenylphenyl)methylamino]-9-propan-2-yl-purin-2-yl]amino]butan-1-ol, Cyclin-K, Cyclin-dependent kinase 12, ... | | 著者 | Kozicka, Z, Kempf, G, Petzold, G, Thoma, N.H. | | 登録日 | 2022-11-30 | | 公開日 | 2023-09-13 | | 最終更新日 | 2024-01-03 | | 実験手法 | X-RAY DIFFRACTION (3.08 Å) | | 主引用文献 | Design principles for cyclin K molecular glue degraders.
Nat.Chem.Biol., 20, 2024
|
|
8BUA
 
 | | Structure of DDB1 bound to 919278-engaged CDK12-cyclin K | | 分子名称: | (2~{R})-~{N}-(1~{H}-benzimidazol-2-yl)-2-(3-oxidanylidene-1~{H}-isoindol-2-yl)propanamide, CITRIC ACID, Cyclin-K, ... | | 著者 | Kozicka, Z, Kempf, G, Petzold, G, Thoma, N.H. | | 登録日 | 2022-11-30 | | 公開日 | 2023-09-13 | | 最終更新日 | 2024-01-03 | | 実験手法 | X-RAY DIFFRACTION (3.193 Å) | | 主引用文献 | Design principles for cyclin K molecular glue degraders.
Nat.Chem.Biol., 20, 2024
|
|
