3L33
 
 | | Human mesotrypsin complexed with amyloid precursor protein inhibitor(APPI) | | 分子名称: | Amyloid beta A4 protein, CALCIUM ION, FORMIC ACID, ... | | 著者 | Salameh, M.A, Soares, A.S, Radisky, E.S. | | 登録日 | 2009-12-16 | | 公開日 | 2010-09-22 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (2.48 Å) | | 主引用文献 | Determinants of affinity and proteolytic stability in interactions of Kunitz family protease inhibitors with mesotrypsin.
J.Biol.Chem., 285, 2010
|
|
7JYC
 
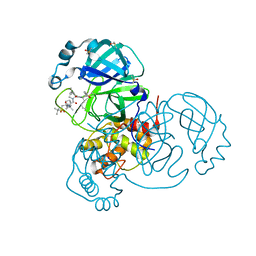 | | Crystal Structure of SARS-CoV-2 Main Protease (3CLpro/Mpro) in Complex with Covalent Inhibitor Narlaprevir | | 分子名称: | (1R,2S,5S)-3-[N-({1-[(tert-butylsulfonyl)methyl]cyclohexyl}carbamoyl)-3-methyl-L-valyl]-N-{(1S)-1-[(1R)-2-(cyclopropylamino)-1-hydroxy-2-oxoethyl]pentyl}-6,6-dimethyl-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase, DIMETHYL SULFOXIDE, ... | | 著者 | Andi, B, Kumaran, D, Kreitler, D.F, Soares, A.S, Shi, W, Jakoncic, J, Fuchs, M.R, Keereetaweep, J, Shanklin, J, McSweeney, S. | | 登録日 | 2020-08-30 | | 公開日 | 2020-09-09 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.79 Å) | | 主引用文献 | Hepatitis C virus NS3/4A inhibitors and other drug-like compounds as covalent binders of SARS-CoV-2 main protease.
Sci Rep, 12, 2022
|
|
7K3T
 
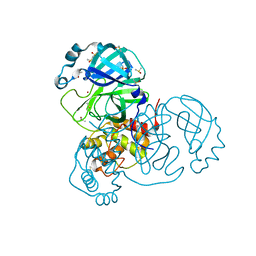 | | Crystal Structure of SARS-CoV-2 Main Protease (3CLpro/Mpro) at 1.2 A Resolution and a Possible Capture of Zinc Binding Intermediate | | 分子名称: | 3C-like proteinase, DIMETHYL SULFOXIDE, ZINC ION | | 著者 | Andi, B, Kumaran, D, Kreitler, D.F, Soares, A.S, Shi, W, Jakoncic, J, Fuchs, M.R, Keereetaweep, J, Shanklin, J, McSweeney, S. | | 登録日 | 2020-09-13 | | 公開日 | 2020-09-23 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.2 Å) | | 主引用文献 | Hepatitis C virus NS3/4A inhibitors and other drug-like compounds as covalent binders of SARS-CoV-2 main protease.
Sci Rep, 12, 2022
|
|
7K6D
 
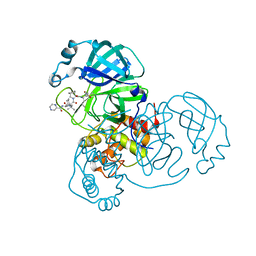 | | SARS-CoV-2 Main Protease Co-Crystal Structure with Telaprevir Determined from Crystals Grown with 40 nL Acoustically Ejected Mpro Droplets at 1.48 A Resolution (Cryo-protected) | | 分子名称: | (1S,3aR,6aS)-2-[(2S)-2-({(2S)-2-cyclohexyl-2-[(pyrazin-2-ylcarbonyl)amino]acetyl}amino)-3,3-dimethylbutanoyl]-N-[(2R,3S)-1-(cyclopropylamino)-2-hydroxy-1-oxohexan-3-yl]octahydrocyclopenta[c]pyrrole-1-carboxamide, 3C-like proteinase, DIMETHYL SULFOXIDE | | 著者 | Kreitler, D.F, Andi, B, Kumaran, D, Soares, A.S, Shi, W, Jakoncic, J, Fuchs, M.R, Keereetaweep, J, Shanklin, J, McSweeney, S. | | 登録日 | 2020-09-19 | | 公開日 | 2020-09-30 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.48 Å) | | 主引用文献 | Hepatitis C virus NS3/4A inhibitors and other drug-like compounds as covalent binders of SARS-CoV-2 main protease.
Sci Rep, 12, 2022
|
|
7K6E
 
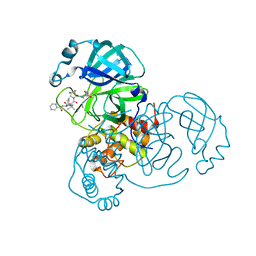 | | SARS-CoV-2 Main Protease Co-Crystal Structure with Telaprevir Determined from Crystals Grown with 40 nL Acoustically Ejected Mpro Droplets at 1.63 A Resolution (Direct Vitrification) | | 分子名称: | (1S,3aR,6aS)-2-[(2S)-2-({(2S)-2-cyclohexyl-2-[(pyrazin-2-ylcarbonyl)amino]acetyl}amino)-3,3-dimethylbutanoyl]-N-[(2R,3S)-1-(cyclopropylamino)-2-hydroxy-1-oxohexan-3-yl]octahydrocyclopenta[c]pyrrole-1-carboxamide, 3C-like proteinase, DIMETHYL SULFOXIDE | | 著者 | Kreitler, D.F, Andi, B, Kumaran, D, Soares, A.S, Shi, W, Jakoncic, J, Fuchs, M.R, Keereetaweep, J, Shanklin, J, McSweeney, S. | | 登録日 | 2020-09-19 | | 公開日 | 2020-09-30 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.63 Å) | | 主引用文献 | Hepatitis C virus NS3/4A inhibitors and other drug-like compounds as covalent binders of SARS-CoV-2 main protease.
Sci Rep, 12, 2022
|
|
7K40
 
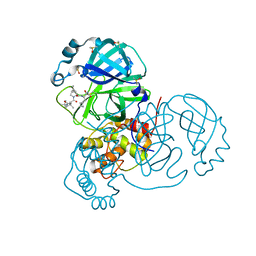 | | Crystal Structure of SARS-CoV-2 Main Protease (3CLpro/Mpro) in Complex with Covalent Inhibitor Boceprevir at 1.35 A Resolution | | 分子名称: | 3C-like proteinase, DIMETHYL SULFOXIDE, boceprevir (bound form) | | 著者 | Kumaran, D, Andi, B, Kreitler, D.F, Soares, A.S, Shi, W, Jakoncic, J, Fuchs, M.R, Keereetaweep, J, Shanklin, J, McSweeney, S. | | 登録日 | 2020-09-14 | | 公開日 | 2020-09-23 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.35 Å) | | 主引用文献 | Hepatitis C virus NS3/4A inhibitors and other drug-like compounds as covalent binders of SARS-CoV-2 main protease.
Sci Rep, 12, 2022
|
|
7MNG
 
 | | Crystal Structure of SARS-CoV-2 Main Protease (3CLpro/Mpro) in Complex with Covalent Inhibitor VBY-825 (Partial Occupancy) | | 分子名称: | (2R,3S)-N-cyclopropyl-3-{[(2R)-3-(cyclopropylmethanesulfonyl)-2-{[(1S)-2,2,2-trifluoro-1-(4-fluorophenyl)ethyl]amino}propanoyl]amino}-2-hydroxypentanamide (non-preferred name), 3C-like proteinase, DIMETHYL SULFOXIDE | | 著者 | Andi, B, Kumaran, D, Soares, A.S, Kreitler, D.F, Shi, W, Jakoncic, J, Fuchs, M.R, Keereetaweep, J, Shanklin, J, McSweeney, S. | | 登録日 | 2021-04-30 | | 公開日 | 2021-05-12 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Hepatitis C virus NS3/4A inhibitors and other drug-like compounds as covalent binders of SARS-CoV-2 main protease.
Sci Rep, 12, 2022
|
|
7MRR
 
 | | Crystal Structure of SARS-CoV-2 Main Protease (3CLpro/Mpro) in Complex with Covalent Inhibitor Leupeptin | | 分子名称: | 3C-like proteinase, DIMETHYL SULFOXIDE, LEUPEPTIN | | 著者 | Andi, B, Kumaran, D, Soares, A.S, Kreitler, D.F, Shi, W, Jakoncic, J, Fuchs, M.R, Keereetaweep, J, Shanklin, J, McSweeney, S. | | 登録日 | 2021-05-08 | | 公開日 | 2021-05-19 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.32 Å) | | 主引用文献 | Hepatitis C virus NS3/4A inhibitors and other drug-like compounds as covalent binders of SARS-CoV-2 main protease.
Sci Rep, 12, 2022
|
|
7KU0
 
 | | Data clustering and dynamics of chymotrypsinogen cluster 138 (yellow) structure | | 分子名称: | Chymotrypsinogen A, SULFATE ION | | 著者 | Nguyen, T, Phan, K.L, Kreitler, D.F, Andrews, L.C, Gabelli, S.B, Kozakov, D, Jakoncic, J, Sweet, R.M, Soares, A.S, Bernstein, H.J. | | 登録日 | 2020-11-24 | | 公開日 | 2021-03-10 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.02 Å) | | 主引用文献 | A simple technique to classify diffraction data from dynamic proteins according to individual polymorphs.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7KU2
 
 | | Data clustering and dynamics of chymotrypsinogen clulster 140 (structure) | | 分子名称: | Chymotrypsinogen A, SULFATE ION | | 著者 | Nguyen, T, Phan, K.L, Kreitler, D.F, Andrews, L.C, Gabelli, S.B, Kozakov, D, Jakoncic, J, Sweet, R.M, Soares, A.S, Bernstein, H.J. | | 登録日 | 2020-11-24 | | 公開日 | 2021-03-10 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.185 Å) | | 主引用文献 | A simple technique to classify diffraction data from dynamic proteins according to individual polymorphs.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7KU1
 
 | | Data clustering and dynamics of chymotrypsinogen cluster 139 (green) structure | | 分子名称: | Chymotrypsinogen A, SULFATE ION | | 著者 | Nguyen, T, Phan, K.L, Kreitler, D.F, Andrews, L.C, Gabelli, S.B, Kozakov, D, Jakoncic, J, Sweet, R.M, Soares, A.S, Bernstein, H.J. | | 登録日 | 2020-11-24 | | 公開日 | 2021-03-10 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.39 Å) | | 主引用文献 | A simple technique to classify diffraction data from dynamic proteins according to individual polymorphs.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7KU3
 
 | | Data clustering and dynamics of chymotrypsinogen cluster 141 (cyan) structure | | 分子名称: | Chymotrypsinogen A, SULFATE ION | | 著者 | Nguyen, T, Phan, K.L, Kreitler, D.F, Andrews, L.C, Gabelli, S.B, Kozakov, D, Jakoncic, J, Sweet, R.M, Soares, A.S, Bernstein, H.J. | | 登録日 | 2020-11-24 | | 公開日 | 2021-03-10 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | A simple technique to classify diffraction data from dynamic proteins according to individual polymorphs.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7KTZ
 
 | | Data clustering and dynamics of chymotrypsinogen cluster 131 (purple) structure | | 分子名称: | Chymotrypsinogen A, SULFATE ION | | 著者 | Nguyen, T, Phan, K.L, Kreitler, D.F, Andrews, L.C, Gabelli, S.B, Kozakov, D, Jakoncic, J, Sweet, R.M, Soares, A.S, Bernstein, H.J. | | 登録日 | 2020-11-24 | | 公開日 | 2021-03-10 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | A simple technique to classify diffraction data from dynamic proteins according to individual polymorphs.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7KTY
 
 | | Data clustering and dynamics of chymotrypsinogen average structure | | 分子名称: | Chymotrypsinogen A, SULFATE ION | | 著者 | Nguyen, T, Phan, K.L, Kreitler, D.F, Andrews, L.C, Gabelli, S.B, Kozakov, D, Jakoncic, J, Shi, W, Sweet, R.M, Soares, A.S, Bernstein, H.J. | | 登録日 | 2020-11-24 | | 公開日 | 2021-03-10 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | A simple technique to classify diffraction data from dynamic proteins according to individual polymorphs.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
5JBT
 
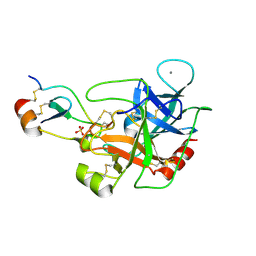 | | Mesotrypsin in complex with cleaved amyloid precursor like protein 2 inhibitor (APLP2) | | 分子名称: | Amyloid-like protein 2, CALCIUM ION, PRSS3 protein, ... | | 著者 | Kayode, O, Wang, R, Pendlebury, D, Soares, A, Radisky, E.S. | | 登録日 | 2016-04-13 | | 公開日 | 2016-11-09 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | An Acrobatic Substrate Metamorphosis Reveals a Requirement for Substrate Conformational Dynamics in Trypsin Proteolysis.
J. Biol. Chem., 291, 2016
|
|
4IQS
 
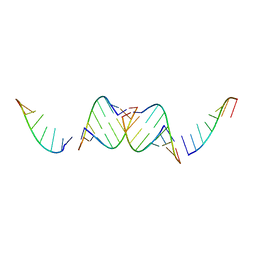 | |
8G83
 
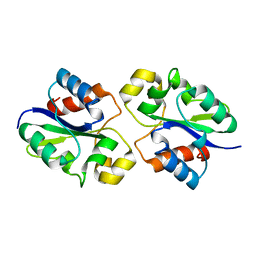 | | Structure of NAD+ consuming protein Acinetobacter baumannii TIR domain | | 分子名称: | NAD(+) hydrolase AbTIR | | 著者 | Klontz, E.H, Wang, Y, Glendening, G, Carr, J, Tsibouris, T, Buddula, S, Nallar, S, Soares, A, Snyder, G.A. | | 登録日 | 2023-02-17 | | 公開日 | 2023-10-11 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (3.03 Å) | | 主引用文献 | The structure of NAD + consuming protein Acinetobacter baumannii TIR domain shows unique kinetics and conformations.
J.Biol.Chem., 299, 2023
|
|
2CGO
 
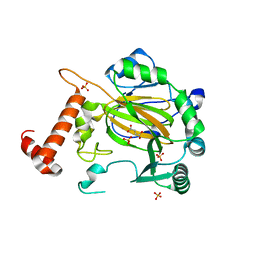 | | FACTOR INHIBITING HIF-1 ALPHA with fumarate | | 分子名称: | FE (III) ION, FUMARIC ACID, HYPOXIA-INDUCIBLE FACTOR 1 ALPHA INHIBITOR, ... | | 著者 | McDonough, M.A, Clifton, I.J, Schofield, C.J. | | 登録日 | 2006-03-09 | | 公開日 | 2006-12-13 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Structural and Mechanistic Studies on the Inhibition of the Hypoxia-Inducible Transcription Factor Hydroxylases by Tricarboxylic Acid Cycle Intermediates.
J.Biol.Chem., 282, 2007
|
|
2CGN
 
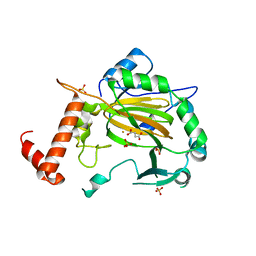 | | FACTOR INHIBITING HIF-1 ALPHA with succinate | | 分子名称: | FE (III) ION, HYPOXIA-INDUCIBLE FACTOR 1 ALPHA INHIBITOR, SUCCINIC ACID, ... | | 著者 | McDonough, M.A, Clifton, I.J, Schofield, C.J. | | 登録日 | 2006-03-09 | | 公開日 | 2006-12-13 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Structural and Mechanistic Studies on the Inhibition of the Hypoxia-Inducible Transcription Factor Hydroxylases by Tricarboxylic Acid Cycle Intermediates.
J.Biol.Chem., 282, 2007
|
|
4EKN
 
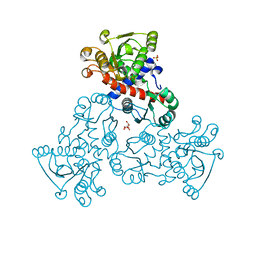 | |
6NCI
 
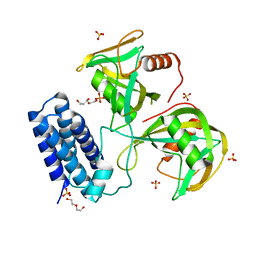 | | Crystal structure of CDP-Chase: Vector data collection | | 分子名称: | D-ribose, DI(HYDROXYETHYL)ETHER, PHOSPHATE ION, ... | | 著者 | Miller, M.S, Shi, W, Gabelli, S.B. | | 登録日 | 2018-12-11 | | 公開日 | 2019-02-06 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.08 Å) | | 主引用文献 | Getting the Most Out of Your Crystals: Data Collection at the New High-Flux, Microfocus MX Beamlines at NSLS-II.
Molecules, 24, 2019
|
|
6NCT
 
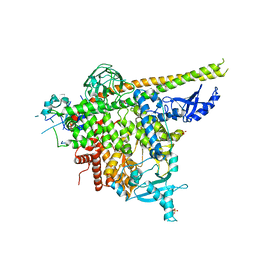 | | Structure of p110alpha/niSH2 - vector data collection | | 分子名称: | Phosphatidylinositol 3-kinase regulatory subunit alpha, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform, SULFATE ION, ... | | 著者 | Miller, M.S, Maheshwari, S, Amzel, L.M, Gabelli, S.B. | | 登録日 | 2018-12-12 | | 公開日 | 2019-02-06 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (3.35 Å) | | 主引用文献 | Getting the Most Out of Your Crystals: Data Collection at the New High-Flux, Microfocus MX Beamlines at NSLS-II.
Molecules, 24, 2019
|
|
6NCK
 
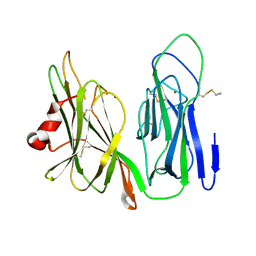 | | Crystal structure of H108A peptidylglycine alpha-hydroxylating monooxygenase (PHM) | | 分子名称: | COPPER (II) ION, NICKEL (II) ION, Peptidyl-glycine alpha-amidating monooxygenase | | 著者 | Miller, M.S, Maheshwari, S, Gabelli, S.B. | | 登録日 | 2018-12-11 | | 公開日 | 2019-02-06 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Getting the Most Out of Your Crystals: Data Collection at the New High-Flux, Microfocus MX Beamlines at NSLS-II.
Molecules, 24, 2019
|
|
6NCH
 
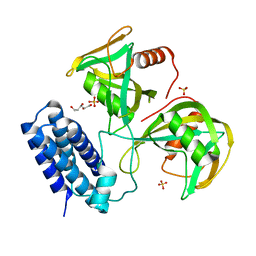 | | Crystal structure of CDP-Chase: Raster data collection | | 分子名称: | D-ribose, PHOSPHATE ION, Phosphohydrolase (MutT/nudix family protein), ... | | 著者 | Miller, M.S, Shi, W, Gabelli, S.B. | | 登録日 | 2018-12-11 | | 公開日 | 2019-02-06 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Getting the Most Out of Your Crystals: Data Collection at the New High-Flux, Microfocus MX Beamlines at NSLS-II.
Molecules, 24, 2019
|
|
3LTR
 
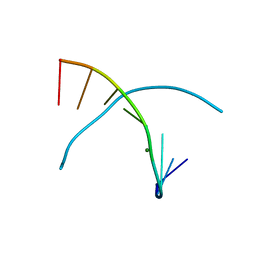 | | 5-OMe-dU containing DNA 8mer | | 分子名称: | 5'-D(*GP*(UMS)P*GP*(T5O)P*AP*CP*AP*C)-3', MAGNESIUM ION | | 著者 | Sheng, J, Hassan, A.E.A, Zhang, W, Gan, J, Huang, Z. | | 登録日 | 2010-02-16 | | 公開日 | 2010-03-09 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.3 Å) | | 主引用文献 | Hydrogen bond formation between the naturally modified nucleobase and phosphate backbone.
Nucleic Acids Res., 40, 2012
|
|
