7SJR
 
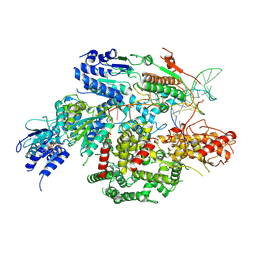 | | Cryo-EM structure of AdnA-AdnB(W325A) in complex with DNA and AMPPNP | | 分子名称: | DNA (70-MER), DNA helicase, IRON/SULFUR CLUSTER, ... | | 著者 | Wang, J, Warren, G.M, Shuman, S, Patel, D.J. | | 登録日 | 2021-10-18 | | 公開日 | 2021-12-22 | | 最終更新日 | 2024-06-05 | | 実験手法 | ELECTRON MICROSCOPY (3.8 Å) | | 主引用文献 | Structure-activity relationships at a nucleobase-stacking tryptophan required for chemomechanical coupling in the DNA resecting motor-nuclease AdnAB.
Nucleic Acids Res., 50, 2022
|
|
8T8F
 
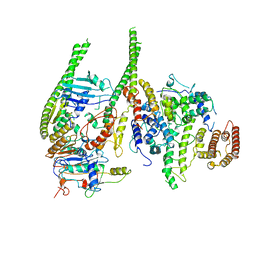 | | Smc5/6 8mer | | 分子名称: | DNA repair protein KRE29, Non-structural maintenance of chromosome element 4, Non-structural maintenance of chromosome element 5, ... | | 著者 | Yu, Y, Patel, D.J. | | 登録日 | 2023-06-22 | | 公開日 | 2023-11-15 | | 実験手法 | ELECTRON MICROSCOPY (4.8 Å) | | 主引用文献 | Molecular basis for Nse5-6 mediated regulation of Smc5/6 functions.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8T8E
 
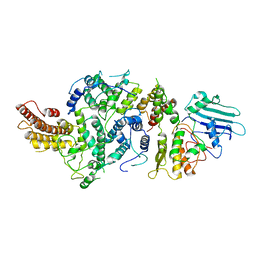 | | cryoEM structure of Smc5/6 5mer | | 分子名称: | DNA repair protein KRE29, Non-structural maintenance of chromosome element 5, Structural maintenance of chromosomes protein 6 | | 著者 | Yu, Y, Patel, D.J. | | 登録日 | 2023-06-22 | | 公開日 | 2023-11-15 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | Molecular basis for Nse5-6 mediated regulation of Smc5/6 functions.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8T66
 
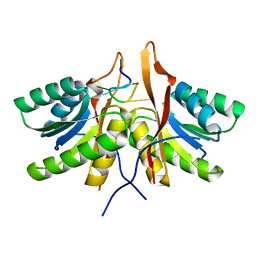 | | cA6 bound Cam1 | | 分子名称: | Cam1, RNA (5'-R(P*AP*AP*AP*AP*A)-3') | | 著者 | Yu, Y, Patel, D.J. | | 登録日 | 2023-06-15 | | 公開日 | 2024-01-10 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | The CRISPR effector Cam1 mediates membrane depolarization for phage defence.
Nature, 625, 2024
|
|
8T64
 
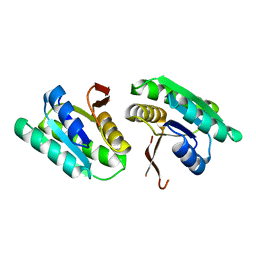 | | Apo Cam1(42-206) | | 分子名称: | Cam1 | | 著者 | Yu, Y, Patel, D.J. | | 登録日 | 2023-06-15 | | 公開日 | 2024-01-10 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.25 Å) | | 主引用文献 | The CRISPR effector Cam1 mediates membrane depolarization for phage defence.
Nature, 625, 2024
|
|
8T65
 
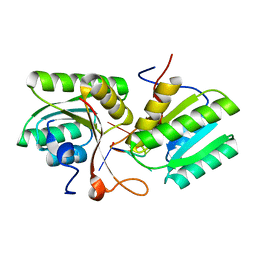 | | cA4 bound Cam1 | | 分子名称: | Cam1, RNA (5'-R(P*AP*AP*AP*A)-3') | | 著者 | Yu, Y, Patel, D.J. | | 登録日 | 2023-06-15 | | 公開日 | 2024-01-10 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.09 Å) | | 主引用文献 | The CRISPR effector Cam1 mediates membrane depolarization for phage defence.
Nature, 625, 2024
|
|
5IX2
 
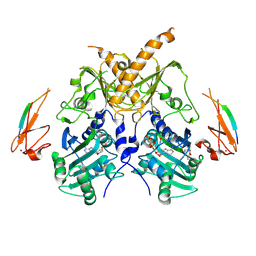 | | Crystal structure of mouse Morc3 ATPase-CW cassette in complex with AMPPNP and unmodified H3 peptide | | 分子名称: | MAGNESIUM ION, MORC family CW-type zinc finger protein 3, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | 著者 | Li, S, Du, J, Patel, D.J. | | 登録日 | 2016-03-23 | | 公開日 | 2016-08-17 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Mouse MORC3 is a GHKL ATPase that localizes to H3K4me3 marked chromatin
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5IW5
 
 | |
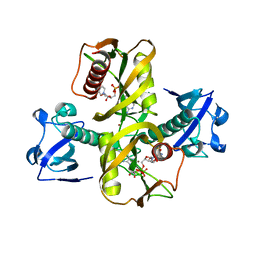
5IW4
 
 | |
6EDB
 
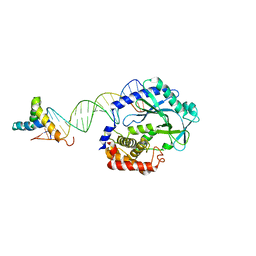 | | Crystal structure of SRY.hcGAS-21bp dsDNA complex | | 分子名称: | DNA (5'-D(*CP*GP*AP*TP*CP*CP*GP*GP*GP*AP*TP*CP*TP*AP*AP*AP*CP*AP*AP*TP*G)-3'), DNA (5'-D(*GP*CP*AP*TP*TP*GP*TP*TP*TP*AP*GP*AP*TP*CP*CP*CP*GP*GP*AP*TP*C)-3'), Sex-determining region Y protein,Cyclic GMP-AMP synthase, ... | | 著者 | Xie, W, Lama, L, Adura, C, Glickman, J.F, Tuschl, T, Patel, D.J. | | 登録日 | 2018-08-09 | | 公開日 | 2019-05-29 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (3.209 Å) | | 主引用文献 | Human cGAS catalytic domain has an additional DNA-binding interface that enhances enzymatic activity and liquid-phase condensation.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
3UIH
 
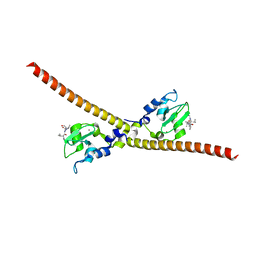 | |
3UIG
 
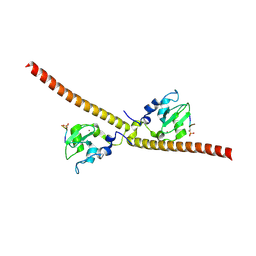 | |
3VRS
 
 | | Crystal structure of fluoride riboswitch, soaked in Mn2+ | | 分子名称: | FLUORIDE ION, Fluoride riboswitch, MANGANESE (II) ION, ... | | 著者 | Ren, A.M, Rajashankar, K.R, Patel, D.J. | | 登録日 | 2012-04-13 | | 公開日 | 2012-05-09 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.603 Å) | | 主引用文献 | Fluoride ion encapsulation by Mg2+ ions and phosphates in a fluoride riboswitch.
Nature, 486, 2012
|
|
3UII
 
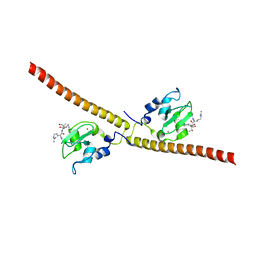 | |
484D
 
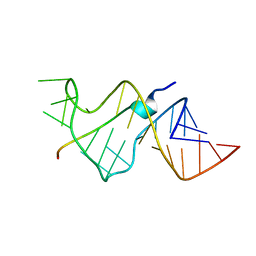 | | SOLUTION STRUCTURE OF HIV-1 REV PEPTIDE-RNA APTAMER COMPLEX | | 分子名称: | BASIC REV PEPTIDE, RNA APTAMER | | 著者 | Ye, X, Gorin, A.A, Frederick, R, Hu, W, Majumdar, A, Xu, W, Mclendon, G, Ellington, A, Patel, D.J. | | 登録日 | 1999-08-02 | | 公開日 | 1999-10-14 | | 最終更新日 | 2023-12-27 | | 実験手法 | SOLUTION NMR | | 主引用文献 | RNA architecture dictates the conformations of a bound peptide.
Chem.Biol., 6, 1999
|
|
149D
 
 | |
1SLP
 
 | | FIRST STEM LOOP OF THE SL1 RNA FROM CAENORHABDITIS ELEGANS, NMR, 16 STRUCTURES | | 分子名称: | RNA (5'-R(*UP*UP*AP*CP*CP*CP*AP*AP*GP*UP*UP*UP*GP*AP*GP*GP*UP*AP*A)-3') | | 著者 | Greenbaum, N.L, Radhakrishnan, I, Patel, D.J, Hirsh, D. | | 登録日 | 1996-05-24 | | 公開日 | 1997-04-21 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of the donor site of a trans-splicing RNA.
Structure, 4, 1996
|
|
1SLO
 
 | | FIRST STEM LOOP OF THE SL1 RNA FROM CAENORHABDITIS ELEGANS, NMR, MINIMIZED AVERAGE STRUCTURE | | 分子名称: | RNA (5'-R(*UP*UP*AP*CP*CP*CP*AP*AP*GP*UP*UP*UP*GP*AP*GP*GP*UP*AP*A)-3') | | 著者 | Greenbaum, N.L, Radhakrishnan, I, Patel, D.J, Hirsh, D. | | 登録日 | 1996-05-24 | | 公開日 | 1996-12-07 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of the donor site of a trans-splicing RNA.
Structure, 4, 1996
|
|
1FJB
 
 | | NMR Study of an 11-Mer DNA Duplex Containing 7,8-Dihydro-8-Oxoadenine (AOXO) Opposite Thymine | | 分子名称: | DNA (5'-D(*CP*GP*TP*AP*CP*(A38)P*CP*AP*TP*GP*C)-3'), DNA (5'-D(*GP*CP*AP*TP*GP*TP*GP*TP*AP*CP*G)-3') | | 著者 | Chen, H, Johnson, F, Grollman, A.P, Patel, D.J. | | 登録日 | 1995-12-15 | | 公開日 | 1996-06-10 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structural Studies of the Ionizing Radiation Adduct 7,8-Dihydro-8-Oxoadenine (A Oxo) Positioned Opposite Thymine and Guanine in DNA Duplexes
MAGN.RESON.CHEM., 34, 1996
|
|
1FMN
 
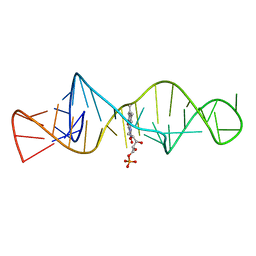 | | SOLUTION STRUCTURE OF FMN-RNA APTAMER COMPLEX, NMR, 5 STRUCTURES | | 分子名称: | FLAVIN MONONUCLEOTIDE, RNA (5'-R(*GP*GP*CP*GP*UP*GP*UP*AP*GP*GP *AP*UP*AP*UP*GP*CP*UP*UP*CP*GP*GP*CP*AP*GP*AP*AP*GP *GP*AP*CP*AP*CP*GP*CP*C)-3') | | 著者 | Fan, P, Suri, A.K, Fiala, R, Live, D, Patel, D.J. | | 登録日 | 1995-12-04 | | 公開日 | 1996-07-11 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Molecular recognition in the FMN-RNA aptamer complex.
J.Mol.Biol., 258, 1996
|
|
1FJA
 
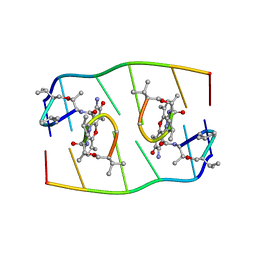 | |
1G70
 
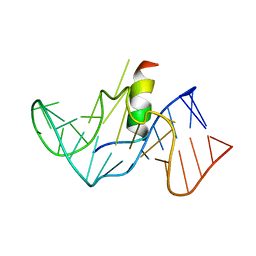 | | COMPLEX OF HIV-1 RRE-IIB RNA WITH RSG-1.2 PEPTIDE | | 分子名称: | HIV-1 RRE-IIB 32 NUCLEOTIDE RNA, RSG-1.2 PEPTIDE | | 著者 | Gosser, Y, Hermann, T, Majumdar, A, Hu, W, Frederick, R, Jiang, F, Xu, W, Patel, D.J. | | 登録日 | 2000-11-08 | | 公開日 | 2001-02-07 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Peptide-triggered conformational switch in HIV-1 RRE RNA complexes.
Nat.Struct.Biol., 8, 2001
|
|
7R78
 
 | |
7R77
 
 | |
7LHL
 
 | |
