8WUT
 
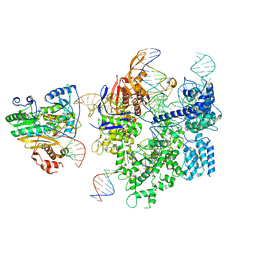 | | SpCas9-MMLV RT-pegRNA-target DNA complex (initiation) | | 分子名称: | CRISPR-associated endonuclease Cas9/Csn1, DNA (5'-D(*TP*GP*AP*TP*GP*GP*CP*AP*GP*AP*GP*TP*AP*CP*TP*AP*G)-3'), DNA (51-MER), ... | | 著者 | Shuto, Y, Nakagawa, R, Hoki, M, Omura, S.N, Hirano, H, Itoh, Y, Nureki, O. | | 登録日 | 2023-10-21 | | 公開日 | 2024-06-05 | | 最終更新日 | 2024-07-17 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | Structural basis for pegRNA-guided reverse transcription by a prime editor.
Nature, 631, 2024
|
|
8WUS
 
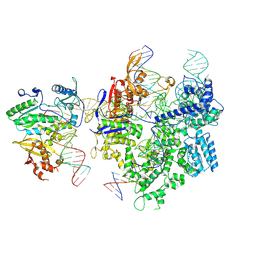 | | SpCas9-MMLV RT-pegRNA-target DNA complex (termination) | | 分子名称: | CRISPR-associated endonuclease Cas9/Csn1, DNA (40-MER), DNA (5'-D(*TP*GP*AP*TP*GP*GP*CP*AP*GP*AP*GP*TP*AP*CP*TP*AP*G)-3'), ... | | 著者 | Shuto, Y, Nakagawa, R, Hoki, M, Omura, S.N, Hirano, H, Itoh, Y, Nureki, O. | | 登録日 | 2023-10-21 | | 公開日 | 2024-06-05 | | 最終更新日 | 2024-07-17 | | 実験手法 | ELECTRON MICROSCOPY (2.9 Å) | | 主引用文献 | Structural basis for pegRNA-guided reverse transcription by a prime editor.
Nature, 631, 2024
|
|
8WUV
 
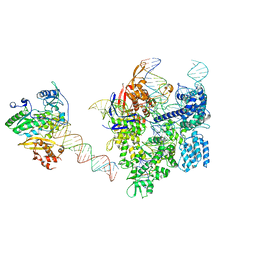 | | SpCas9-MMLV RT-pegRNA-target DNA complex (elongation 16-nt) | | 分子名称: | CRISPR-associated endonuclease Cas9/Csn1, DNA (5'-D(*TP*GP*AP*TP*GP*GP*CP*AP*GP*AP*GP*TP*AP*CP*TP*AP*G)-3'), DNA (50-MER), ... | | 著者 | Shuto, Y, Nakagawa, R, Hoki, M, Omura, S.N, Hirano, H, Itoh, Y, Nureki, O. | | 登録日 | 2023-10-21 | | 公開日 | 2024-06-05 | | 最終更新日 | 2024-07-17 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | Structural basis for pegRNA-guided reverse transcription by a prime editor.
Nature, 631, 2024
|
|
8WUU
 
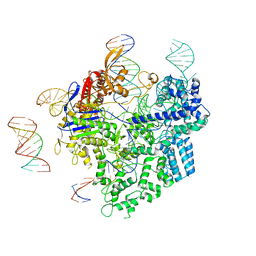 | | SpCas9-pegRNA-target DNA complex (pre-initiation) | | 分子名称: | CRISPR-associated endonuclease Cas9/Csn1, DNA (34-MER), DNA (5'-D(*TP*GP*AP*TP*GP*GP*CP*AP*GP*AP*GP*TP*AP*CP*TP*AP*G)-3'), ... | | 著者 | Shuto, Y, Nakagawa, R, Hoki, M, Omura, S.N, Hirano, H, Itoh, Y, Nureki, O. | | 登録日 | 2023-10-21 | | 公開日 | 2024-06-05 | | 最終更新日 | 2024-07-17 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Structural basis for pegRNA-guided reverse transcription by a prime editor.
Nature, 631, 2024
|
|
8YGJ
 
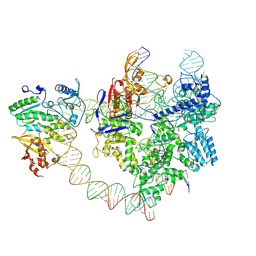 | | SpCas9-MMLV RT-pegRNA-target DNA complex (elongation 28-nt) | | 分子名称: | CRISPR-associated endonuclease Cas9/Csn1, DNA (5'-D(P*TP*GP*AP*TP*GP*GP*CP*AP*GP*AP*GP*TP*AP*CP*TP*AP*G)-3'), DNA (51-MER), ... | | 著者 | Shuto, Y, Nakagawa, R, Hoki, M, Omura, S.N, Hirano, H, Itoh, Y, Nureki, O. | | 登録日 | 2024-02-26 | | 公開日 | 2024-06-05 | | 最終更新日 | 2024-07-17 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Structural basis for pegRNA-guided reverse transcription by a prime editor.
Nature, 631, 2024
|
|
8YFY
 
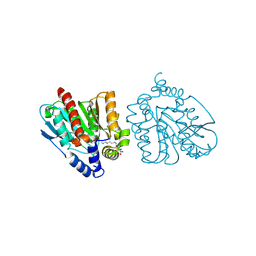 | | CRYSTAL STRUCTURE OF THE EST1 H274D MUTANT AT PH 4.2 | | 分子名称: | Carboxylesterase, octyl beta-D-glucopyranoside | | 著者 | Unno, H, Oshima, Y, Nishino, T, Nakayama, T, Kusunoki, M. | | 登録日 | 2024-02-26 | | 公開日 | 2024-07-10 | | 実験手法 | X-RAY DIFFRACTION (1.45 Å) | | 主引用文献 | Lowering pH optimum of activity of SshEstI, a slightly alkaliphilic archaeal esterase of the hormone-sensitive lipase family.
J.Biosci.Bioeng., 2024
|
|
8YFZ
 
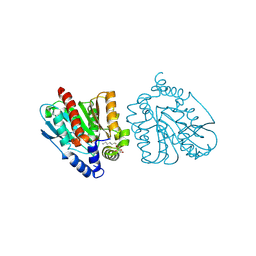 | | CRYSTAL STRUCTURE OF THE EST1 H274E MUTANT AT PH 4.2 | | 分子名称: | Carboxylesterase, octyl beta-D-glucopyranoside | | 著者 | Unno, H, Oshima, Y, Nishino, T, Nakayama, T, Kusunoki, M. | | 登録日 | 2024-02-26 | | 公開日 | 2024-07-10 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Lowering pH optimum of activity of SshEstI, a slightly alkaliphilic archaeal esterase of the hormone-sensitive lipase family.
J.Biosci.Bioeng., 2024
|
|
1WRB
 
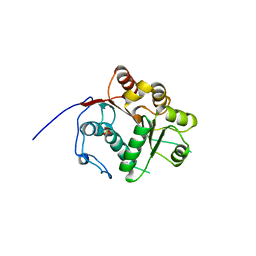 | | Crystal structure of the N-terminal RecA-like domain of DjVLGB, a pranarian Vasa-like RNA helicase | | 分子名称: | DjVLGB, SULFATE ION | | 著者 | Kurimoto, K, Muto, Y, Obayashi, N, Terada, T, Shirouzu, M, Yabuki, T, Aoki, M, Seki, E, Matsuda, T, Kigawa, T, Okumura, H, Tanaka, A, Shibata, N, Kashikawa, M, Agata, K, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | 登録日 | 2004-10-14 | | 公開日 | 2005-04-12 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Crystal structure of the N-terminal RecA-like domain of a DEAD-box RNA helicase, the Dugesia japonica vasa-like gene B protein
J.Struct.Biol., 150, 2005
|
|
1UG2
 
 | | Solution Structure of Mouse Hypothetical Gene (2610100B20Rik) Product Homologous to Myb DNA-binding Domain | | 分子名称: | 2610100B20Rik gene product | | 著者 | Zhao, C, Kigawa, T, Tochio, N, Koshiba, S, Inoue, M, Shirouzu, M, Terada, T, Yabuki, T, Aoki, M, Seki, E, Matsuda, T, Tanaka, A, Osanai, T, Arakawa, T, Carninci, P, Kawai, J, Hayashizaki, Y, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | 登録日 | 2003-06-11 | | 公開日 | 2004-06-22 | | 最終更新日 | 2023-12-27 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution Structure of Mouse Hypothetical Gene (2610100B20Rik) Product Homologous to Myb DNA-binding Domain
To be Published
|
|
1WMZ
 
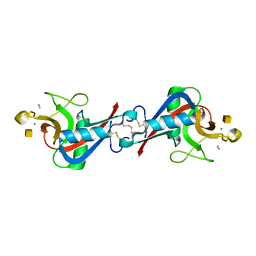 | | Crystal Structure of C-type Lectin CEL-I complexed with N-acetyl-D-galactosamine | | 分子名称: | 2-acetamido-2-deoxy-alpha-D-galactopyranose, 2-acetamido-2-deoxy-beta-D-galactopyranose, CALCIUM ION, ... | | 著者 | Sugawara, H, Kusunoki, M, Kurisu, G, Fujimoto, T, Aoyagi, H, Hatakeyama, T. | | 登録日 | 2004-07-22 | | 公開日 | 2004-09-07 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Characteristic Recognition of N-Acetylgalactosamine by an Invertebrate C-type Lectin, CEL-I, Revealed by X-ray Crystallographic Analysis
J.Biol.Chem., 279, 2004
|
|
1WMY
 
 | | Crystal Structure of C-type Lectin CEL-I from Cucumaria echinata | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, lectin CEL-I, ... | | 著者 | Sugawara, H, Kusunoki, M, Kurisu, G, Fujimoto, T, Aoyagi, H, Hatakeyama, T. | | 登録日 | 2004-07-22 | | 公開日 | 2004-09-07 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Characteristic Recognition of N-Acetylgalactosamine by an Invertebrate C-type Lectin, CEL-I, Revealed by X-ray Crystallographic Analysis
J.Biol.Chem., 279, 2004
|
|
1UF8
 
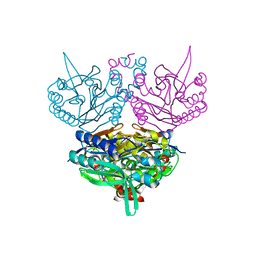 | | Crystal structure of C171A/V236A Mutant of N-carbamyl-D-amino acid amidohydrolase complexed with N-carbamyl-D-Phenylalanine | | 分子名称: | D-[(AMINO)CARBONYL]PHENYLALANINE, N-carbamyl-D-amino acid amidohydrolase | | 著者 | Hashimoto, H, Aoki, M, Shimizu, T, Nakai, T, Morikawa, H, Ikenaka, Y, Takahashi, S, Sato, M. | | 登録日 | 2003-05-26 | | 公開日 | 2004-06-08 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Crystal structure of C171A/V236A Mutant of N-carbamyl-D-amino acid amidohydrolase
To be published
|
|
1UF4
 
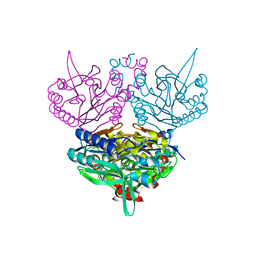 | | Crystal structure of C171A/V236A Mutant of N-carbamyl-D-amino acid amidohydrolase | | 分子名称: | N-carbamyl-D-amino acid amidohydrolase | | 著者 | Hashimoto, H, Aoki, M, Shimizu, T, Nakai, T, Morikawa, H, Ikenaka, Y, Takahashi, S, Sato, M. | | 登録日 | 2003-05-23 | | 公開日 | 2004-06-08 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2.15 Å) | | 主引用文献 | Crystal structure of C171A/V236A Mutant of N-carbamyl-D-amino acid
To be published
|
|
1UF7
 
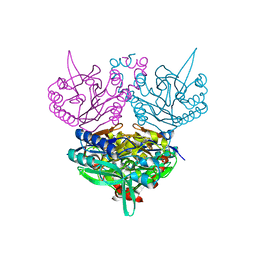 | | Crystal structure of C171A/V236A Mutant of N-carbamyl-D-amino acid amidohydrolase complexed with N-carbamyl-D-valine | | 分子名称: | 3-METHYL-2-UREIDO-BUTYRIC ACID, N-carbamyl-D-amino acid amidohydrolase | | 著者 | Hashimoto, H, Aoki, M, Shimizu, T, Nakai, T, Morikawa, H, Ikenaka, Y, Takahashi, S, Sato, M. | | 登録日 | 2003-05-26 | | 公開日 | 2004-06-08 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Crystal structure of C171A/V236A Mutant of N-carbamyl-D-amino acid amidohydrolase
To be published
|
|
1UF5
 
 | | Crystal structure of C171A/V236A Mutant of N-carbamyl-D-amino acid amidohydrolase complexed with N-carbamyl-D-methionine | | 分子名称: | 1,2-ETHANEDIOL, 4-METHYLSULFANYL-2-UREIDO-BUTYRIC ACID, N-carbamyl-D-amino acid amidohydrolase | | 著者 | Hashimoto, H, Aoki, M, Shimizu, T, Nakai, T, Morikawa, H, Ikenaka, Y, Takahashi, S, Sato, M. | | 登録日 | 2003-05-23 | | 公開日 | 2004-06-08 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Crystal structure of C171A/V236A mutant of N-carbamyl-D-amino acid amidohydrolase
To be published
|
|
1WRI
 
 | | Crystal Structure of Ferredoxin isoform II from E. arvense | | 分子名称: | BENZAMIDINE, FE2/S2 (INORGANIC) CLUSTER, Ferredoxin II | | 著者 | Kurisu, G, Nishiyama, D, Kusunoki, M, Fujikawa, S, Katoh, M, Hanke, G.T, Hase, T, Teshima, K. | | 登録日 | 2004-10-18 | | 公開日 | 2004-11-02 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (1.2 Å) | | 主引用文献 | A structural basis of Equisetum arvense ferredoxin isoform II producing an alternative electron transfer with ferredoxin-NADP+ reductase.
J.Biol.Chem., 280, 2005
|
|
3ASK
 
 | | Structure of UHRF1 in complex with histone tail | | 分子名称: | E3 ubiquitin-protein ligase UHRF1, Histone H3.3, ZINC ION | | 著者 | Arita, K, Sugita, K, Unoki, M, Hamamoto, R, Sekiyama, N, Tochio, H, Ariyoshi, M, Shirakawa, M. | | 登録日 | 2010-12-16 | | 公開日 | 2012-01-25 | | 最終更新日 | 2013-06-05 | | 実験手法 | X-RAY DIFFRACTION (2.904 Å) | | 主引用文献 | Recognition of modification status on a histone H3 tail by linked histone reader modules of the epigenetic regulator UHRF1
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
5GHL
 
 | | Crystal structure Analysis of the starch-binding domain of glucoamylase from Aspergillus niger | | 分子名称: | GLYCEROL, Glucoamylase, SULFATE ION | | 著者 | Miyake, H, Suyama, Y, Muraki, N, Kusunoki, M, Tanaka, A. | | 登録日 | 2016-06-20 | | 公開日 | 2017-10-18 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Crystal structure of the starch-binding domain of glucoamylase from Aspergillus niger.
Acta Crystallogr.,Sect.F, 73, 2017
|
|
3ASL
 
 | | Structure of UHRF1 in complex with histone tail | | 分子名称: | 1,2-ETHANEDIOL, E3 ubiquitin-protein ligase UHRF1, Histone H3.3, ... | | 著者 | Arita, K, Sugita, K, Unoki, M, Hamamoto, R, Sekiyama, N, Tochio, H, Ariyoshi, M, Shirakawa, M. | | 登録日 | 2010-12-16 | | 公開日 | 2012-01-25 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (1.41 Å) | | 主引用文献 | Recognition of modification status on a histone H3 tail by linked histone reader modules of the epigenetic regulator UHRF1
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3WYE
 
 | | Crystal Structure of chimeric engineered (2S,3S)-butanediol dehydrogenase complexed with NAD+ | | 分子名称: | Diacetyl reductase [(S)-acetoin forming],L-2,3-butanediol dehydrogenase,Diacetyl reductase [(S)-acetoin forming],L-2,3-butanediol dehydrogenase,Diacetyl reductase [(S)-acetoin forming],L-2,3-butanediol dehydrogenase,Diacetyl reductase [(S)-acetoin forming], NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | 著者 | Shimegi, T, Oyama, T, Kusunoki, M, Ui, S. | | 登録日 | 2014-08-26 | | 公開日 | 2015-08-26 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (1.58 Å) | | 主引用文献 | Crystal Structure of chimeric engineered (2S,3S)-butanediol dehydrogenase complexed with NAD+
To be Published
|
|
3A28
 
 | | Crystal structure of L-2,3-butanediol dehydrogenase | | 分子名称: | BETA-MERCAPTOETHANOL, L-2.3-butanediol dehydrogenase, MAGNESIUM ION, ... | | 著者 | Otagiri, M, Kurisu, G, Ui, S, Kusunoki, M. | | 登録日 | 2009-05-02 | | 公開日 | 2009-12-15 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structural basis for chiral substrate recognition by two 2,3-butanediol dehydrogenases
Febs Lett., 584, 2010
|
|
6MCS
 
 | | X-ray crystal structure of wild type HIV-1 protease in complex with GRL-003 | | 分子名称: | (3S,3aR,5R,7aS,8S)-hexahydro-4H-3,5-methanofuro[2,3-b]pyran-8-yl [(2S,3R)-4-[{[2-(cyclopropylamino)-1,3-benzothiazol-6-yl]sulfonyl}(2-methylpropyl)amino]-1-(4-fluorophenyl)-3-hydroxybutan-2-yl]carbamate, Protease | | 著者 | Bulut, H, Hayashi, H, Hattori, S.I, Aoki, M, Das, D, Ghosh, A.K, Mitsuya, H. | | 登録日 | 2018-09-02 | | 公開日 | 2019-04-24 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.52 Å) | | 主引用文献 | Halogen Bond Interactions of Novel HIV-1 Protease Inhibitors (PI) (GRL-001-15 and GRL-003-15) with the Flap of Protease Are Critical for Their Potent Activity against Wild-Type HIV-1 and Multi-PI-Resistant Variants.
Antimicrob.Agents Chemother., 63, 2019
|
|
6MCR
 
 | | X-ray crystal structure of wild type HIV-1 protease in complex with GRL-001 | | 分子名称: | (3S,3aR,5R,7aS,8S)-hexahydro-4H-3,5-methanofuro[2,3-b]pyran-8-yl [(2S,3R)-4-[{[2-(cyclopropylamino)-1,3-benzothiazol-6-yl]sulfonyl}(2-methylpropyl)amino]-1-(3-fluorophenyl)-3-hydroxybutan-2-yl]carbamate, 1,2-ETHANEDIOL, Protease | | 著者 | Bulut, H, Hayashi, H, Hattori, S.I, Aoki, M, Das, D, Ghosh, A.K, Mitsuya, H. | | 登録日 | 2018-09-02 | | 公開日 | 2019-04-24 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.48 Å) | | 主引用文献 | Halogen Bond Interactions of Novel HIV-1 Protease Inhibitors (PI) (GRL-001-15 and GRL-003-15) with the Flap of Protease Are Critical for Their Potent Activity against Wild-Type HIV-1 and Multi-PI-Resistant Variants.
Antimicrob.Agents Chemother., 63, 2019
|
|
2E7P
 
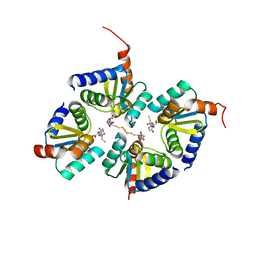 | | Crystal structure of the holo form of glutaredoxin C1 from populus tremula x tremuloides | | 分子名称: | FE2/S2 (INORGANIC) CLUSTER, GLUTATHIONE, Glutaredoxin | | 著者 | Unno, H, Takahashi, T, Kawakami, T, Aimoto, S, Hase, T, Kusunoki, M, Rouhier, N, Jacquot, J.P. | | 登録日 | 2007-01-12 | | 公開日 | 2007-09-25 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Functional, structural, and spectroscopic characterization of a glutathione-ligated [2Fe-2S] cluster in poplar glutaredoxin C1
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
1X1N
 
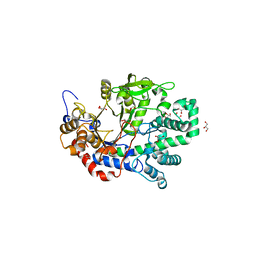 | | Structure determination and refinement at 1.8 A resolution of Disproportionating Enzyme from Potato | | 分子名称: | 4-alpha-glucanotransferase, CALCIUM ION, GLYCEROL | | 著者 | Imamura, K, Matsuura, T, Takaha, T, Fujii, K, Nakagawa, A, Kusunoki, M, Nitta, Y. | | 登録日 | 2005-04-08 | | 公開日 | 2006-04-18 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Structure determination and refinement at 1.8 A resolution of Disproportionating Enzyme from Potato
to be published
|
|
