4DX8
 
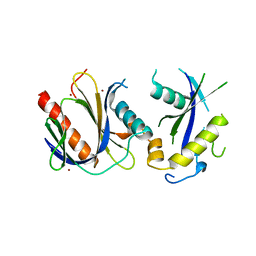 | | ICAP1 in complex with KRIT1 N-terminus | | 分子名称: | BROMIDE ION, Integrin beta-1-binding protein 1, Krev interaction trapped protein 1 | | 著者 | Liu, W, Draheim, K, Zhang, R, Calderwood, D.A, Boggon, T.J. | | 登録日 | 2012-02-27 | | 公開日 | 2013-01-09 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.54 Å) | | 主引用文献 | Mechanism for KRIT1 Release of ICAP1-Mediated Suppression of Integrin Activation.
Mol.Cell, 49, 2013
|
|
4KGG
 
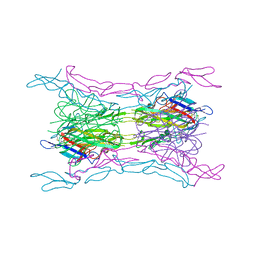 | | Crystal structure of light mutant2 and dcr3 complex | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, MAGNESIUM ION, Tumor necrosis factor ligand superfamily member 14, ... | | 著者 | Liu, W, Bonanno, J.B, Zhan, C, Kumar, P.R, Toro, R, Nathenson, S.G, Almo, S.C, Atoms-to-Animals: The Immune Function Network (IFN), New York Structural Genomics Research Consortium (NYSGRC) | | 登録日 | 2013-04-29 | | 公開日 | 2013-08-07 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.78 Å) | | 主引用文献 | Mechanistic basis for functional promiscuity in the TNF and TNF receptor superfamilies: structure of the LIGHT:DcR3 assembly.
Structure, 22, 2014
|
|
4KG8
 
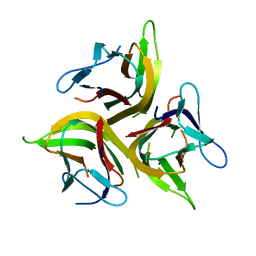 | | Crystal structure of light mutant | | 分子名称: | Tumor necrosis factor ligand superfamily member 14 | | 著者 | Liu, W, Zhan, C, Kumar, P.R, Bonanno, J.B, Nathenson, S.G, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC), Atoms-to-Animals: The Immune Function Network (IFN) | | 登録日 | 2013-04-28 | | 公開日 | 2013-08-07 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.25 Å) | | 主引用文献 | Mechanistic basis for functional promiscuity in the TNF and TNF receptor superfamilies: structure of the LIGHT:DcR3 assembly.
Structure, 22, 2014
|
|
4KGQ
 
 | | Crystal structure of a human light loop mutant in complex with dcr3 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, MAGNESIUM ION, Tumor necrosis factor ligand superfamily member 14, ... | | 著者 | Liu, W, Zhan, C, Bonanno, J.B, Sampathkumar, P, Toro, R, Nathenson, S.G, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC), Atoms-to-Animals: The Immune Function Network (IFN) | | 登録日 | 2013-04-29 | | 公開日 | 2013-07-10 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.27 Å) | | 主引用文献 | Mechanistic basis for functional promiscuity in the TNF and TNF receptor superfamilies: structure of the LIGHT:DcR3 assembly.
Structure, 22, 2014
|
|
3ULR
 
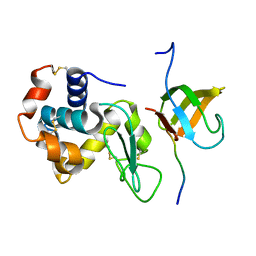 | | Lysozyme contamination facilitates crystallization of a hetero-trimericCortactin:Arg:Lysozyme complex | | 分子名称: | Abelson tyrosine-protein kinase 2, Lysozyme C, Src substrate cortactin | | 著者 | Liu, W, MacGrath, S, Koleske, A.J, Boggon, T.J. | | 登録日 | 2011-11-11 | | 公開日 | 2012-01-11 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | Lysozyme contamination facilitates crystallization of a heterotrimeric cortactin-Arg-lysozyme complex.
Acta Crystallogr.,Sect.F, 68, 2012
|
|
8J1I
 
 | |
8HZT
 
 | |
8HZQ
 
 | |
8WOT
 
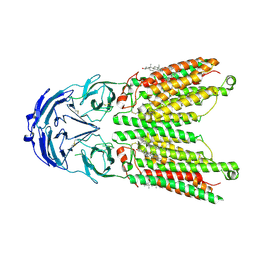 | | Cryo-EM structure of human SIDT1 protein with C2 symmetry at low pH | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL, ... | | 著者 | Liu, W, Tang, M, Wang, J, Zhang, X, Wu, S, Ru, H. | | 登録日 | 2023-10-07 | | 公開日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (3.18 Å) | | 主引用文献 | Structural insights into cholesterol transport and hydrolase activity of a putative human RNA transport protein SIDT1.
Cell Discov, 10, 2024
|
|
8WOQ
 
 | | Cryo-EM structure of human SIDT1 protein with C1 symmetry at neutral pH | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | 著者 | Liu, W, Tang, M, Wang, J, Zhang, X, Wu, S, Ru, H. | | 登録日 | 2023-10-07 | | 公開日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (2.85 Å) | | 主引用文献 | Structural insights into cholesterol transport and hydrolase activity of a putative human RNA transport protein SIDT1.
Cell Discov, 10, 2024
|
|
8WOS
 
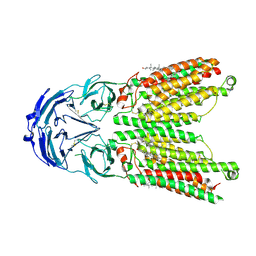 | | Cryo-EM structure of human SIDT1 protein with C1 symmetry at low pH | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL, ... | | 著者 | Liu, W, Tang, M, Wang, J, Zhang, X, Wu, S, Ru, H. | | 登録日 | 2023-10-07 | | 公開日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (3.37 Å) | | 主引用文献 | Structural insights into cholesterol transport and hydrolase activity of a putative human RNA transport protein SIDT1.
Cell Discov, 10, 2024
|
|
8WOR
 
 | | Cryo-EM structure of human SIDT1 protein with C2 symmetry at neutral pH | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | 著者 | Liu, W, Tang, M, Wang, J, Zhang, X, Wu, S, Ru, H. | | 登録日 | 2023-10-07 | | 公開日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (2.66 Å) | | 主引用文献 | Structural insights into cholesterol transport and hydrolase activity of a putative human RNA transport protein SIDT1.
Cell Discov, 10, 2024
|
|
5YF8
 
 | |
5YF6
 
 | |
5YF7
 
 | |
5YF5
 
 | |
6AE6
 
 | |
6AE4
 
 | |
6AE7
 
 | |
6AE5
 
 | |
6JJ0
 
 | |
6K63
 
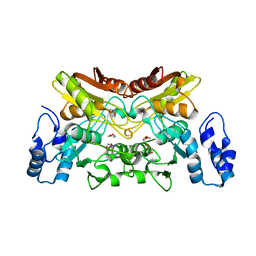 | | The crystal structure of cytidine deaminase from Klebsiella pneumoniae | | 分子名称: | 1,4-DIETHYLENE DIOXIDE, Cytidine deaminase, ZINC ION | | 著者 | Liu, W, Shang, F, Lan, J, Chen, Y, Wang, L, Xu, Y. | | 登録日 | 2019-06-01 | | 公開日 | 2019-07-31 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.073 Å) | | 主引用文献 | Biochemical and structural analysis of the Klebsiella pneumoniae cytidine deaminase CDA.
Biochem.Biophys.Res.Commun., 519, 2019
|
|
3R0H
 
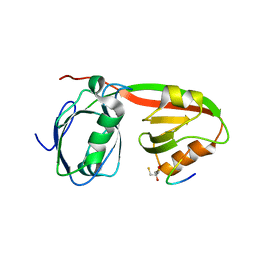 | | Structure of INAD PDZ45 in complex with NG2 peptide | | 分子名称: | (2S,3S)-1,4-DIMERCAPTOBUTANE-2,3-DIOL, 2,3-DIHYDROXY-1,4-DITHIOBUTANE, Inactivation-no-after-potential D protein, ... | | 著者 | Wei, Z, Liu, W, Zhang, M. | | 登録日 | 2011-03-08 | | 公開日 | 2011-11-30 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | The INAD scaffold is a dynamic, redox-regulated modulator of signaling in the Drosophila eye
Cell(Cambridge,Mass.), 145, 2011
|
|
3D6V
 
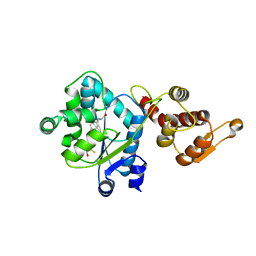 | | Crystal structure of 4-(trifluoromethyldiazirinyl)phenylalanyl-tRNA synthetase | | 分子名称: | 4-(2,2,2-TRIFLUOROETHYL)-L-PHENYLALANINE, BETA-MERCAPTOETHANOL, Tyrosyl-tRNA synthetase | | 著者 | Liu, W, Tippmann, E, Mack, A.V, Schultz, P.G. | | 登録日 | 2008-05-20 | | 公開日 | 2008-05-27 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | A genetically encoded diazirine photocrosslinker in Escherichia coli
ChemBioChem, 8, 2007
|
|
3D6U
 
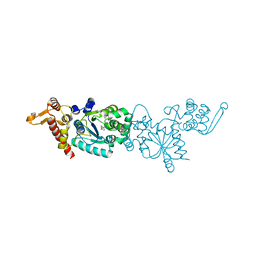 | | Crystal structure of 4-(trifluoromethyldiazirinyl)phenylalanyl-tRNA synthetase | | 分子名称: | 4-[3-(TRIFLUOROMETHYL)DIAZIRIDIN-3-YL]-L-PHENYLALANINE, BETA-MERCAPTOETHANOL, Tyrosyl-tRNA synthetase | | 著者 | Liu, W, Tippmann, E, Mack, A.V, Schultz, P.G. | | 登録日 | 2008-05-20 | | 公開日 | 2008-05-27 | | 最終更新日 | 2021-10-20 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | A genetically encoded diazirine photocrosslinker in Escherichia coli
ChemBioChem, 8, 2007
|
|
