6OUG
 
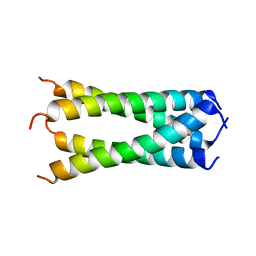 | | Structure of drug-resistant V27A mutant of the influenza M2 proton channel bound to spiroadamantyl amine inhibitor, TM + cytosolic helix construct | | 分子名称: | (1r,1'S,3'S,5'S,7'S)-spiro[cyclohexane-1,2'-tricyclo[3.3.1.1~3,7~]decan]-4-amine, Matrix protein 2 | | 著者 | Thomaston, J.L, Liu, L, DeGrado, W.F. | | 登録日 | 2019-05-04 | | 公開日 | 2020-01-15 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (3.01 Å) | | 主引用文献 | X-ray Crystal Structures of the Influenza M2 Proton Channel Drug-Resistant V27A Mutant Bound to a Spiro-Adamantyl Amine Inhibitor Reveal the Mechanism of Adamantane Resistance.
Biochemistry, 59, 2020
|
|
6NV1
 
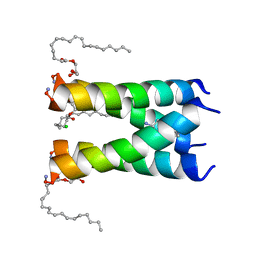 | | Structure of drug-resistant V27A mutant of the influenza M2 proton channel bound to spiroadamantyl amine inhibitor | | 分子名称: | (1r,1'S,3'S,5'S,7'S)-spiro[cyclohexane-1,2'-tricyclo[3.3.1.1~3,7~]decan]-4-amine, (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, CHLORIDE ION, ... | | 著者 | Thomaston, J.L, Liu, L, DeGrado, W.F. | | 登録日 | 2019-02-04 | | 公開日 | 2020-01-15 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | X-ray Crystal Structures of the Influenza M2 Proton Channel Drug-Resistant V27A Mutant Bound to a Spiro-Adamantyl Amine Inhibitor Reveal the Mechanism of Adamantane Resistance.
Biochemistry, 59, 2020
|
|
1PWO
 
 | |
6GEL
 
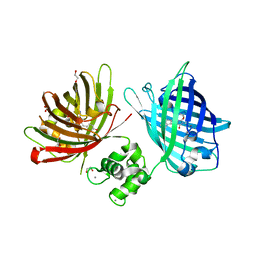 | | The structure of TWITCH-2B | | 分子名称: | CALCIUM ION, FORMIC ACID, GLYCEROL, ... | | 著者 | Trigo Mourino, P, Paulat, M, Thestrup, T, Griesbeck, O, Griesinger, C, Becker, S. | | 登録日 | 2018-04-26 | | 公開日 | 2019-08-21 | | 最終更新日 | 2019-09-11 | | 実験手法 | X-RAY DIFFRACTION (2.51 Å) | | 主引用文献 | Dynamic tuning of FRET in a green fluorescent protein biosensor.
Sci Adv, 5, 2019
|
|
6GEZ
 
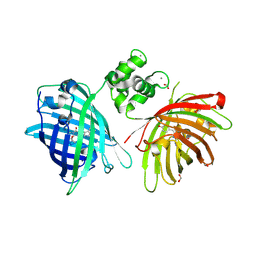 | | THE STRUCTURE OF TWITCH-2B N532F | | 分子名称: | CALCIUM ION, FORMIC ACID, Green fluorescent protein,Optimized Ratiometric Calcium Sensor,Green fluorescent protein,Green fluorescent protein | | 著者 | Trigo Mourino, P, Paulat, M, Thestrup, T, Griesbeck, O, Griesinger, C, Becker, S. | | 登録日 | 2018-04-27 | | 公開日 | 2019-08-21 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (2.47 Å) | | 主引用文献 | Dynamic tuning of FRET in a green fluorescent protein biosensor.
Sci Adv, 5, 2019
|
|
5KBR
 
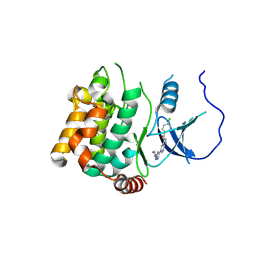 | | Pak1 in complex with 7-azaindole inhibitor | | 分子名称: | (4-chlorophenyl)-[5-(1-piperidin-4-ylpyrazol-4-yl)-1~{H}-pyrrolo[2,3-b]pyridin-3-yl]methanone, Serine/threonine-protein kinase PAK 1 | | 著者 | Ferguson, A. | | 登録日 | 2016-06-03 | | 公開日 | 2016-09-28 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.36 Å) | | 主引用文献 | Optimization of Highly Kinase Selective Bis-anilino Pyrimidine PAK1 Inhibitors.
ACS Med Chem Lett, 7, 2016
|
|
5KBQ
 
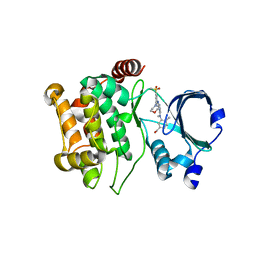 | | Pak1 in complex with bis-anilino pyrimidine inhibitor | | 分子名称: | Serine/threonine-protein kinase PAK 1, [4-methyl-3-[methyl-[2-[(3-methylsulfonyl-5-morpholin-4-yl-phenyl)amino]pyrimidin-4-yl]amino]phenyl]methanol | | 著者 | Ferguson, A. | | 登録日 | 2016-06-03 | | 公開日 | 2016-09-28 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.58 Å) | | 主引用文献 | Optimization of Highly Kinase Selective Bis-anilino Pyrimidine PAK1 Inhibitors.
ACS Med Chem Lett, 7, 2016
|
|
5VTG
 
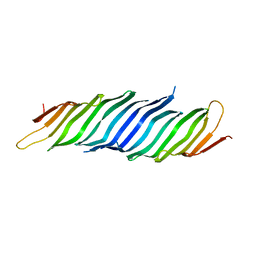 | |
1P7O
 
 | |
1OZY
 
 | |
7ZH2
 
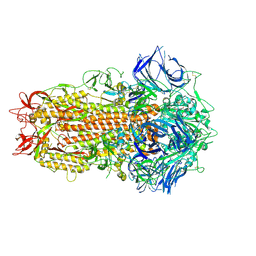 | | SARS CoV Spike protein, Closed C1 conformation | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, LINOLEIC ACID, ... | | 著者 | Toelzer, C, Gupta, K, Yadav, S.K.N, Buzas, D, Borucu, U, Schaffitzel, C, Berger, I. | | 登録日 | 2022-04-05 | | 公開日 | 2023-02-15 | | 実験手法 | ELECTRON MICROSCOPY (2.71 Å) | | 主引用文献 | The free fatty acid-binding pocket is a conserved hallmark in pathogenic beta-coronavirus spike proteins from SARS-CoV to Omicron.
Sci Adv, 8, 2022
|
|
7ZH5
 
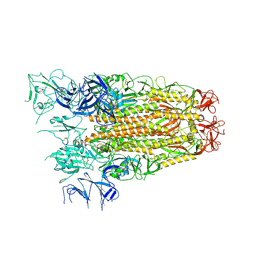 | | SARS CoV Spike protein, Open conformation | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein,Fibritin | | 著者 | Toelzer, C, Gupta, K, Yadav, S.K.N, Buzas, D, Borucu, U, Schaffitzel, C, Berger, I. | | 登録日 | 2022-04-05 | | 公開日 | 2023-02-15 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | The free fatty acid-binding pocket is a conserved hallmark in pathogenic beta-coronavirus spike proteins from SARS-CoV to Omicron.
Sci Adv, 8, 2022
|
|
7ZH1
 
 | | SARS CoV Spike protein, Closed C3 conformation | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, LINOLEIC ACID, ... | | 著者 | Toelzer, C, Gupta, K, Yadav, S.K.N, Buzas, D, Borucu, U, Schaffitzel, C, Berger, I. | | 登録日 | 2022-04-05 | | 公開日 | 2023-02-15 | | 実験手法 | ELECTRON MICROSCOPY (2.48 Å) | | 主引用文献 | The free fatty acid-binding pocket is a conserved hallmark in pathogenic beta-coronavirus spike proteins from SARS-CoV to Omicron.
Sci Adv, 8, 2022
|
|
2M97
 
 | |
1Y6O
 
 | | Crystal structure of disulfide engineered porcine pancreatic phospholipase A2 to group-X isozyme in complex with inhibitor MJ33 and phosphate ions | | 分子名称: | 1-HEXADECYL-3-TRIFLUOROETHYL-SN-GLYCERO-2-PHOSPHATE METHANE, CALCIUM ION, PHOSPHATE ION, ... | | 著者 | Yu, B.Z, Pan, Y.H, Jassen, M.J.W, Bahnson, B.J, Jain, M.K. | | 登録日 | 2004-12-06 | | 公開日 | 2005-03-22 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Kinetic and structural properties of disulfide engineered phospholipase a(2): insight into the role of disulfide bonding patterns.
Biochemistry, 44, 2005
|
|
1Y6P
 
 | | Crystal structure of disulfide engineered porcine pancratic phospholipase a2 to group-x isozyme | | 分子名称: | CALCIUM ION, CHLORIDE ION, Phospholipase A2, ... | | 著者 | Yu, B.Z, Pan, Y.H, Jassen, M.J.W, Bahnson, B.J, Jain, M.K. | | 登録日 | 2004-12-06 | | 公開日 | 2005-03-22 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (2.25 Å) | | 主引用文献 | Kinetic and structural properties of disulfide engineered phospholipase a(2): insight into the role of disulfide bonding patterns.
Biochemistry, 44, 2005
|
|
7ROV
 
 | |
7JRN
 
 | | Crystal structure of the wild type SARS-CoV-2 papain-like protease (PLPro) with inhibitor GRL0617 | | 分子名称: | 5-amino-2-methyl-N-[(1R)-1-naphthalen-1-ylethyl]benzamide, Non-structural protein 3, SULFATE ION, ... | | 著者 | Sacco, M, Ma, C, Wang, J, Chen, Y. | | 登録日 | 2020-08-12 | | 公開日 | 2020-08-26 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.48 Å) | | 主引用文献 | Discovery of SARS-CoV-2 Papain-like Protease Inhibitors through a Combination of High-Throughput Screening and a FlipGFP-Based Reporter Assay.
Acs Cent.Sci., 7, 2021
|
|
7L51
 
 | |
7L53
 
 | |
7L54
 
 | |
7L55
 
 | |
7M3U
 
 | |
2KNI
 
 | |
1LMM
 
 | | Solution Structure of Psmalmotoxin 1, the First Characterized Specific Blocker of ASIC1a NA+ channel | | 分子名称: | Psalmotoxin 1 | | 著者 | Escoubas, P, Bernard, C, Lazdunski, M, Darbon, H. | | 登録日 | 2002-05-02 | | 公開日 | 2003-11-25 | | 最終更新日 | 2022-02-23 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Recombinant production and solution structure of PcTx1, the specific peptide inhibitor of ASIC1a proton-gated cation channels
Protein Sci., 12, 2003
|
|
