4BN6
 
 | |
3B4O
 
 | | Crystal structure of phenazine biosynthesis protein PhzA/B from Burkholderia cepacia R18194, apo form | | 分子名称: | ACETATE ION, Phenazine biosynthesis protein A/B | | 著者 | Ahuja, E.G, Janning, P, Mentel, M, Graebsch, A, Breinbauer, R, Blankenfeldt, W. | | 登録日 | 2007-10-24 | | 公開日 | 2008-12-30 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | PhzA/B catalyzes the formation of the tricycle in phenazine biosynthesis.
J.Am.Chem.Soc., 130, 2008
|
|
3B4P
 
 | | Crystal structure of phenazine biosynthesis protein PhzA/B from Burkholderia cepacia R18194, complex with 2-(cyclohexylamino)benzoic acid | | 分子名称: | 2-(cyclohexylamino)benzoic acid, ACETATE ION, AZIDE ION, ... | | 著者 | Ahuja, E.G, Janning, P, Mentel, M, Graebsch, A, Breinbauer, R, Blankenfeldt, W. | | 登録日 | 2007-10-24 | | 公開日 | 2008-12-30 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | PhzA/B catalyzes the formation of the tricycle in phenazine biosynthesis.
J.Am.Chem.Soc., 130, 2008
|
|
1SOT
 
 | | Crystal Structure of the DegS stress sensor | | 分子名称: | Protease degS | | 著者 | Wilken, C, Kitzing, K, Kurzbauer, R, Ehrmann, M, Clausen, T. | | 登録日 | 2004-03-15 | | 公開日 | 2004-06-08 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Crystal structure of the DegS stress sensor: How a PDZ domain recognizes misfolded protein and activates a protease
Cell(Cambridge,Mass.), 117, 2004
|
|
1RZM
 
 | | Crystal structure of 3-deoxy-D-arabino-heptulosonate-7-phosphate synthase (DAHPS) from Thermotoga maritima complexed with Cd2+, PEP and E4P | | 分子名称: | CADMIUM ION, ERYTHOSE-4-PHOSPHATE, PHOSPHOENOLPYRUVATE, ... | | 著者 | Shumilin, I.A, Bauerle, R, Wu, J, Woodard, R.W, Kretsinger, R.H. | | 登録日 | 2003-12-24 | | 公開日 | 2004-08-10 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Crystal Structure of the Reaction Complex of 3-Deoxy-d-arabino-heptulosonate-7-phosphate Synthase from Thermotoga maritima Refines the Catalytic Mechanism and Indicates a New Mechanism of Allosteric Regulation.
J.Mol.Biol., 341, 2004
|
|
1DNY
 
 | | SOLUTION STRUCTURE OF PCP, A PROTOTYPE FOR THE PEPTIDYL CARRIER DOMAINS OF MODULAR PEPTIDE SYNTHETASES | | 分子名称: | NON-RIBOSOMAL PEPTIDE SYNTHETASE PEPTIDYL CARRIER PROTEIN | | 著者 | Weber, T, Baumgartner, R, Renner, C, Marahiel, M.A, Holak, T.A. | | 登録日 | 1999-12-17 | | 公開日 | 2000-05-17 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of PCP, a prototype for the peptidyl carrier domains of modular peptide synthetases.
Structure Fold.Des., 8, 2000
|
|
1SOZ
 
 | | Crystal Structure of DegS protease in complex with an activating peptide | | 分子名称: | Protease degS, activating peptide | | 著者 | Wilken, C, Kitzing, K, Kurzbauer, R, Ehrmann, M, Clausen, T. | | 登録日 | 2004-03-16 | | 公開日 | 2004-06-08 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Crystal structure of the DegS stress sensor: How a PDZ domain recognizes misfolded protein and activates a protease
Cell(Cambridge,Mass.), 117, 2004
|
|
1VCW
 
 | | Crystal structure of DegS after backsoaking the activating peptide | | 分子名称: | Protease degS | | 著者 | Wilken, C, Kitzing, K, Kurzbauer, R, Ehrmann, M, Clausen, T. | | 登録日 | 2004-03-16 | | 公開日 | 2004-06-08 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (3.05 Å) | | 主引用文献 | Crystal structure of the DegS stress sensor: How a PDZ domain recognizes misfolded protein and activates a protease.
Cell(Cambridge,Mass.), 117, 2004
|
|
4BN7
 
 | |
4BNB
 
 | |
3DZL
 
 | | Crystal structure of PhzA/B from Burkholderia cepacia R18194 in complex with (R)-3-oxocyclohexanecarboxylic acid | | 分子名称: | (1R)-3-oxocyclohexanecarboxylic acid, Phenazine biosynthesis protein A/B | | 著者 | Ahuja, E.G, Mentel, M, Graebsch, A, Breinbauer, R, Blankenfeldt, W. | | 登録日 | 2008-07-30 | | 公開日 | 2008-12-30 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | PhzA/B Catalyzes the Formation of the Tricycle in Phenazine Biosynthesis.
J.Am.Chem.Soc., 130, 2008
|
|
4BN8
 
 | |
2WON
 
 | | Crystal Structure of UK-453061 bound to HIV-1 Reverse Transcriptase (wild-type). | | 分子名称: | 5-{[3,5-diethyl-1-(2-hydroxyethyl)-1H-pyrazol-4-yl]oxy}benzene-1,3-dicarbonitrile, HIV-1 REVERSE TRANSCRIPTASE | | 著者 | Phillips, C, Irving, S.L, Knoechel, T, Ringrose, H. | | 登録日 | 2009-07-27 | | 公開日 | 2010-08-11 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Lersivirine: A Non-Nucleoside Reverse Transcriptase Inhibitor with Activity Against Drug- Resistant Human Immunodeficiency Virus-1.
Antimicrob.Agents Chemother., 54, 2010
|
|
2WOM
 
 | | Crystal Structure of UK-453061 bound to HIV-1 Reverse Transcriptase (K103N). | | 分子名称: | 5-{[3,5-diethyl-1-(2-hydroxyethyl)-1H-pyrazol-4-yl]oxy}benzene-1,3-dicarbonitrile, HIV-1 REVERSE TRANSCRIPTASE | | 著者 | Phillips, C, Irving, S.L, Knoechel, T, Ringrose, H. | | 登録日 | 2009-07-27 | | 公開日 | 2010-08-11 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (3.2 Å) | | 主引用文献 | Lersivirine, a nonnucleoside reverse transcriptase inhibitor with activity against drug-resistant human immunodeficiency virus type 1.
Antimicrob. Agents Chemother., 54, 2010
|
|
4ZXF
 
 | | Crystal Structure of a Soluble Variant of Monoglyceride Lipase from Saccharomyces Cerevisiae in Complex with a Substrate Analog | | 分子名称: | 1-{3-[(R)-hydroxy(octadecyloxy)phosphoryl]propyl}triaza-1,2-dien-2-ium, Monoglyceride lipase, NITRATE ION, ... | | 著者 | Aschauer, P, Lichtenegger, J, Rengachari, S, Gruber, K, Oberer, M. | | 登録日 | 2015-05-20 | | 公開日 | 2016-05-25 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Crystal structure of the Saccharomyces cerevisiae monoglyceride lipase Yju3p.
Biochim.Biophys.Acta, 1861, 2016
|
|
3CNM
 
 | | Crystal Structure of Phenazine Biosynthesis Protein PhzA/B from Burkholderia cepacia R18194, DHHA complex | | 分子名称: | (2S,3S)-TRANS-2,3-DIHYDRO-3-HYDROXYANTHRANILIC ACID, ACETATE ION, Phenazine biosynthesis protein A/B | | 著者 | Ahuja, E.G, Blankenfeldt, W. | | 登録日 | 2008-03-26 | | 公開日 | 2008-12-30 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | PhzA/B catalyzes the formation of the tricycle in phenazine biosynthesis.
J.Am.Chem.Soc., 130, 2008
|
|
8QMX
 
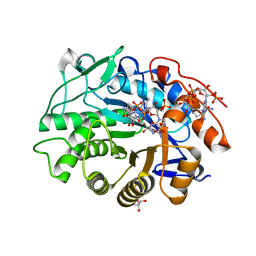 | | OPR3 wildtype in complex with NADPH4 | | 分子名称: | 12-oxophytodienoate reductase 3, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, FLAVIN MONONUCLEOTIDE, ... | | 著者 | Bijelic, A, Macheroux, P, Kerschbaumer, B. | | 登録日 | 2023-09-25 | | 公開日 | 2024-01-17 | | 最終更新日 | 2024-04-10 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | Loop 6 and the beta-hairpin flap are structural hotspots that determine cofactor specificity in the FMN-dependent family of ene-reductases.
Febs J., 291, 2024
|
|
8QN3
 
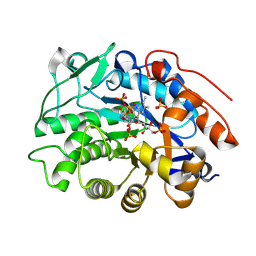 | | OPR3 wildtype in complex with NADH4 | | 分子名称: | 1,4,5,6-Tetrahydronicotinamide adenine dinucleotide, 12-oxophytodienoate reductase 3, FLAVIN MONONUCLEOTIDE, ... | | 著者 | Bijelic, A, Macheroux, P, Keschbaumer, B. | | 登録日 | 2023-09-25 | | 公開日 | 2024-01-17 | | 最終更新日 | 2024-04-10 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Loop 6 and the beta-hairpin flap are structural hotspots that determine cofactor specificity in the FMN-dependent family of ene-reductases.
Febs J., 291, 2024
|
|
2ABZ
 
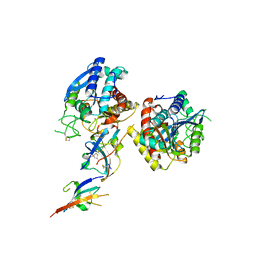 | | Crystal structure of C19A/C43A mutant of leech carboxypeptidase inhibitor in complex with bovine carboxypeptidase A | | 分子名称: | Carboxypeptidase A1, Metallocarboxypeptidase inhibitor, ZINC ION | | 著者 | Arolas, J.L, Popowicz, G.M, Bronsoms, S, Aviles, F.X, Huber, R, Holak, T.A, Ventura, S. | | 登録日 | 2005-07-18 | | 公開日 | 2006-01-31 | | 最終更新日 | 2021-11-10 | | 実験手法 | X-RAY DIFFRACTION (2.16 Å) | | 主引用文献 | Study of a major intermediate in the oxidative folding of leech carboxypeptidase inhibitor: contribution of the fourth disulfide bond
J.Mol.Biol., 352, 2005
|
|
4ZWN
 
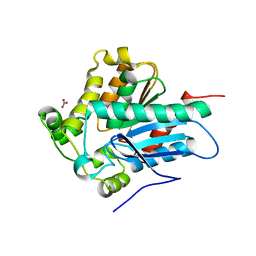 | | Crystal Structure of a Soluble Variant of the Monoglyceride Lipase from Saccharomyces Cerevisiae | | 分子名称: | Monoglyceride lipase, NITRATE ION, SODIUM ION, ... | | 著者 | Aschauer, P, Rengachari, S, Gruber, K, Oberer, M. | | 登録日 | 2015-05-19 | | 公開日 | 2016-04-27 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.491 Å) | | 主引用文献 | Crystal structure of the Saccharomyces cerevisiae monoglyceride lipase Yju3p.
Biochim.Biophys.Acta, 1861, 2016
|
|
5K3Y
 
 | | Crystal structure of AuroraB/INCENP in complex with BI 811283 | | 分子名称: | Aurora kinase B-A, Inner centromere protein A, N-methyl-N-(1-methylpiperidin-4-yl)-4-{[4-({(1R,2S)-2-[(propan-2-yl)carbamoyl]cyclopentyl}amino)-5-(trifluoromethyl)pyrimidin-2-yl]amino}benzamide | | 著者 | Bader, G, Zahn, S.K, Zoephel, A. | | 登録日 | 2016-05-20 | | 公開日 | 2016-08-17 | | 最終更新日 | 2022-12-07 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Pharmacological Profile of BI 847325, an Orally Bioavailable, ATP-Competitive Inhibitor of MEK and Aurora Kinases.
Mol.Cancer Ther., 15, 2016
|
|
7BII
 
 | | Crystal structure of Nematocida HUWE1 | | 分子名称: | E3 ubiquitin-protein ligase HUWE1 | | 著者 | Grabarczyk, D.B, Petrova, O.A, Meinhart, A, Kessler, D, Clausen, T. | | 登録日 | 2021-01-12 | | 公開日 | 2021-07-28 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (3.037 Å) | | 主引用文献 | HUWE1 employs a giant substrate-binding ring to feed and regulate its HECT E3 domain.
Nat.Chem.Biol., 17, 2021
|
|
6QDJ
 
 | | Molecular features of the UNC-45 chaperone critical for binding and folding muscle myosin | | 分子名称: | 1,4-BUTANEDIOL, 2,5,8,11,14,17-HEXAOXANONADECAN-19-OL, ADENOSINE-5'-DIPHOSPHATE, ... | | 著者 | Meinhart, A, Clausen, T, Arnese, R. | | 登録日 | 2019-01-02 | | 公開日 | 2019-10-30 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.884 Å) | | 主引用文献 | Molecular features of the UNC-45 chaperone critical for binding and folding muscle myosin.
Nat Commun, 10, 2019
|
|
5D4W
 
 | |
1ZFI
 
 | | Solution structure of the leech carboxypeptidase inhibitor | | 分子名称: | Metallocarboxypeptidase inhibitor | | 著者 | Arolas, J.L, D'Silva, L, Popowicz, G.M, Aviles, F.X, Holak, T.A, Ventura, S. | | 登録日 | 2005-04-20 | | 公開日 | 2005-09-13 | | 最終更新日 | 2022-03-02 | | 実験手法 | SOLUTION NMR | | 主引用文献 | NMR structural characterization and computational predictions of the major intermediate in oxidative folding of leech carboxypeptidase inhibitor
STRUCTURE, 13, 2005
|
|
