4WJS
 
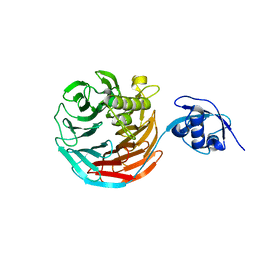 | |
4WJU
 
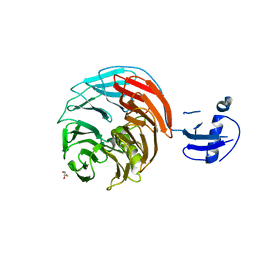 | | Crystal structure of Rsa4 from Saccharomyces cerevisiae | | 分子名称: | GLYCEROL, Ribosome assembly protein 4 | | 著者 | Holdermann, I, Bassler, J, Hurt, E, Sinning, I. | | 登録日 | 2014-10-01 | | 公開日 | 2014-11-19 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | A network of assembly factors is involved in remodeling rRNA elements during preribosome maturation.
J.Cell Biol., 207, 2014
|
|
6RXU
 
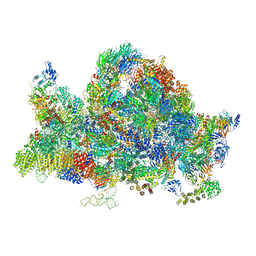 | | Cryo-EM structure of the 90S pre-ribosome (Kre33-Noc4) from Chaetomium thermophilum, state B1 | | 分子名称: | 35S rRNA, 40S ribosomal protein S1, 40S ribosomal protein S11-like protein, ... | | 著者 | Cheng, J, Kellner, N, Griesel, S, Berninghausen, O, Beckmann, R, Hurt, E. | | 登録日 | 2019-06-10 | | 公開日 | 2019-08-14 | | 最終更新日 | 2024-05-22 | | 実験手法 | ELECTRON MICROSCOPY (3.5 Å) | | 主引用文献 | Thermophile 90S Pre-ribosome Structures Reveal the Reverse Order of Co-transcriptional 18S rRNA Subdomain Integration.
Mol.Cell, 75, 2019
|
|
6RXX
 
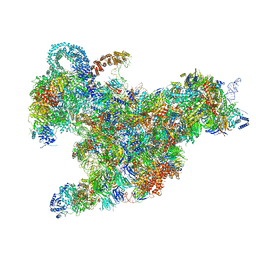 | | Cryo-EM structure of the 90S pre-ribosome (Kre33-Noc4) from Chaetomium thermophilum, state C, Poly-Ala | | 分子名称: | 35S ribosomal RNA, 40S ribosomal protein S1, 40S ribosomal protein S11-like protein, ... | | 著者 | Cheng, J, Kellner, N, Griesel, S, Berninghausen, O, Beckmann, R, Hurt, E. | | 登録日 | 2019-06-10 | | 公開日 | 2019-08-14 | | 最終更新日 | 2024-05-22 | | 実験手法 | ELECTRON MICROSCOPY (7.1 Å) | | 主引用文献 | Thermophile 90S Pre-ribosome Structures Reveal the Reverse Order of Co-transcriptional 18S rRNA Subdomain Integration.
Mol.Cell, 75, 2019
|
|
6RXV
 
 | | Cryo-EM structure of the 90S pre-ribosome (Kre33-Noc4) from Chaetomium thermophilum, state B2 | | 分子名称: | 35S ribosomal RNA, 40S ribosomal protein S1, 40S ribosomal protein S11-like protein, ... | | 著者 | Cheng, J, Kellner, N, Griesel, S, Berninghausen, O, Beckmann, R, Hurt, E. | | 登録日 | 2019-06-10 | | 公開日 | 2019-08-14 | | 最終更新日 | 2019-10-02 | | 実験手法 | ELECTRON MICROSCOPY (4 Å) | | 主引用文献 | Thermophile 90S Pre-ribosome Structures Reveal the Reverse Order of Co-transcriptional 18S rRNA Subdomain Integration.
Mol.Cell, 75, 2019
|
|
6RXY
 
 | | Cryo-EM structure of the 90S pre-ribosome (Kre33-Noc4) from Chaetomium thermophilum, state a | | 分子名称: | 35S rRNA, 40S ribosomal protein S13-like protein, 40S ribosomal protein S14-like protein, ... | | 著者 | Cheng, J, Kellner, N, Griesel, S, Berninghausen, O, Beckmann, R, Hurt, E. | | 登録日 | 2019-06-10 | | 公開日 | 2019-08-14 | | 最終更新日 | 2024-05-22 | | 実験手法 | ELECTRON MICROSCOPY (4.7 Å) | | 主引用文献 | Thermophile 90S Pre-ribosome Structures Reveal the Reverse Order of Co-transcriptional 18S rRNA Subdomain Integration.
Mol.Cell, 75, 2019
|
|
2MVF
 
 | | Structural insight into an essential assembly factor network on the pre-ribosome | | 分子名称: | Uncharacterized protein | | 著者 | Lee, W, Bassler, J, Paternoga, H, Holdermann, I, Thomas, M, Granneman, S, Barrio-Garcia, C, Nyarko, A, Stier, G, Clark, S.A, Schraivogel, D, Kallas, M, Beckmann, R, Tollervey, D, Barbar, E, Sinning, I, Hurt, E. | | 登録日 | 2014-10-02 | | 公開日 | 2014-12-03 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | A network of assembly factors is involved in remodeling rRNA elements during preribosome maturation.
J.Cell Biol., 207, 2014
|
|
5APP
 
 | | Actinobacillus actinomycetemcomitans OMP100 residues 133-198 fused to GCN4 adaptors | | 分子名称: | CHLORIDE ION, General control protein GCN4,GENERAL CONTROL PROTEIN GCN4, OUTER MEMBRANE PROTEIN 100,General control protein GCN4 | | 著者 | Hartmann, M.D, Ridderbusch, O, Lupas, A.N, Hernandez Alvarez, B. | | 登録日 | 2015-09-17 | | 公開日 | 2016-01-27 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | alpha / beta coiled coils.
Elife, 5, 2016
|
|
5APW
 
 | | Sequence MATKDD inserted between GCN4 adaptors - Structure T6 | | 分子名称: | CALCIUM ION, CHLORIDE ION, GENERAL CONTROL PROTEIN GCN4, ... | | 著者 | Hartmann, M.D, Mendler, C.T, Lupas, A.N, Hernandez Alvarez, B. | | 登録日 | 2015-09-17 | | 公開日 | 2016-01-27 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | alpha / beta coiled coils.
Elife, 5, 2016
|
|
5APX
 
 | | Sequence MATKDDIAN inserted between GCN4 adaptors - Structure T9(6) | | 分子名称: | GENERAL CONTROL PROTEIN GCN4, PHOSPHATE ION | | 著者 | Hartmann, M.D, Mendler, C.T, Lupas, A.N, Hernandez Alvarez, B. | | 登録日 | 2015-09-17 | | 公開日 | 2016-01-27 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | alpha / beta coiled coils.
Elife, 5, 2016
|
|
5APT
 
 | |
5APS
 
 | |
5APZ
 
 | | Thermosinus carboxydivorans Nor1 Tcar0761 residues 68-101 and 191-211 fused to GCN4 adaptors | | 分子名称: | GENERAL CONTROL PROTEIN GCN4, NOR1 TCAR0761 | | 著者 | Hartmann, M.D, Deiss, S, Lupas, A.N, Hernandez Alvarez, B. | | 登録日 | 2015-09-17 | | 公開日 | 2016-01-27 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | alpha / beta coiled coils.
Elife, 5, 2016
|
|
5APV
 
 | |
5APU
 
 | | Sequence IANKEDKAD inserted between GCN4 adaptors - Structure A9b black | | 分子名称: | GENERAL CONTROL PROTEIN GCN4, UREA | | 著者 | Hartmann, M.D, Mendler, C.T, Lupas, A.N, Hernandez Alvarez, B. | | 登録日 | 2015-09-17 | | 公開日 | 2016-01-27 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.35 Å) | | 主引用文献 | alpha / beta coiled coils.
Elife, 5, 2016
|
|
5APQ
 
 | |
5APY
 
 | |
7JV2
 
 | | SARS-CoV-2 spike in complex with the S2H13 neutralizing antibody Fab fragment (local refinement of the receptor-binding motif and Fab variable domains) | | 分子名称: | S2H13 Fab heavy chain, S2H13 Fab light chain, Spike glycoprotein | | 著者 | Park, Y.J, Tortorici, M.A, Walls, A.C, Czudnochowski, N, Seattle Structural Genomics Center for Infectious Disease (SSGCID), Snell, G, Veesler, D. | | 登録日 | 2020-08-20 | | 公開日 | 2020-10-14 | | 最終更新日 | 2021-01-27 | | 実験手法 | ELECTRON MICROSCOPY (3.5 Å) | | 主引用文献 | Mapping Neutralizing and Immunodominant Sites on the SARS-CoV-2 Spike Receptor-Binding Domain by Structure-Guided High-Resolution Serology.
Cell, 183, 2020
|
|
7JV4
 
 | | SARS-CoV-2 spike in complex with the S2H13 neutralizing antibody (one RBD open) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, S2H13 Fab heavy chain, ... | | 著者 | Park, Y.J, Tortorici, M.A, Walls, A.C, Czudnochowski, N, Seattle Structural Genomics Center for Infectious Disease (SSGCID), Snell, G, Veesler, D. | | 登録日 | 2020-08-20 | | 公開日 | 2020-10-14 | | 最終更新日 | 2023-01-04 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | Mapping Neutralizing and Immunodominant Sites on the SARS-CoV-2 Spike Receptor-Binding Domain by Structure-Guided High-Resolution Serology.
Cell, 183, 2020
|
|
7JVC
 
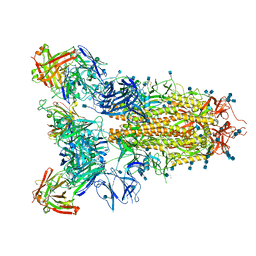 | | SARS-CoV-2 spike in complex with the S2A4 neutralizing antibody Fab fragment | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, S2A4 Fab heavy chain, ... | | 著者 | Park, Y.J, Tortorici, M.A, Walls, A.C, Czudnochowski, N, Seattle Structural Genomics Center for Infectious Disease (SSGCID), Snell, G, Veesler, D. | | 登録日 | 2020-08-20 | | 公開日 | 2020-10-14 | | 最終更新日 | 2021-06-23 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | Mapping Neutralizing and Immunodominant Sites on the SARS-CoV-2 Spike Receptor-Binding Domain by Structure-Guided High-Resolution Serology.
Cell, 183, 2020
|
|
7JW0
 
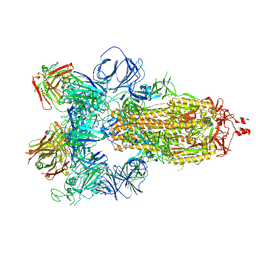 | | SARS-CoV-2 spike in complex with the S304 neutralizing antibody Fab fragment | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, S304 Fab heavy chain, ... | | 著者 | Walls, A.C, Park, Y.J, Tortorici, M.A, Czudnochowski, N, Seattle Structural Genomics Center for Infectious Disease (SSGCID), Snell, G, Veesler, D. | | 登録日 | 2020-08-24 | | 公開日 | 2020-10-14 | | 最終更新日 | 2021-01-27 | | 実験手法 | ELECTRON MICROSCOPY (4.3 Å) | | 主引用文献 | Mapping Neutralizing and Immunodominant Sites on the SARS-CoV-2 Spike Receptor-Binding Domain by Structure-Guided High-Resolution Serology.
Cell, 183, 2020
|
|
7JXC
 
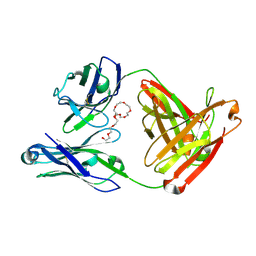 | | Mapping neutralizing and immunodominant sites on the SARS-CoV-2 spike receptor-binding domain by structure-guided high-resolution serology | | 分子名称: | NONAETHYLENE GLYCOL, S2H14 antigen-binding (Fab) fragment | | 著者 | Park, Y.J, Tortorici, M.A, Walls, A.C, Czudnochowski, N, Seattle Structural Genomics Center for Infectious Disease (SSGCID), Snell, G, Veesler, D. | | 登録日 | 2020-08-27 | | 公開日 | 2020-10-14 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.47 Å) | | 主引用文献 | Mapping Neutralizing and Immunodominant Sites on the SARS-CoV-2 Spike Receptor-Binding Domain by Structure-Guided High-Resolution Serology.
Cell, 183, 2020
|
|
7JVA
 
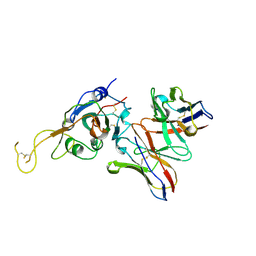 | | SARS-CoV-2 spike in complex with the S2A4 neutralizing antibody Fab fragment (local refinement of the receptor-binding domain and Fab variable domains) | | 分子名称: | S2A4 Fab heavy chain, S2A4 Fab light chain, Spike glycoprotein, ... | | 著者 | Park, Y.J, Tortorici, M.A, Walls, A.C, Czudnochowski, N, Seattle Structural Genomics Center for Infectious Disease (SSGCID), Snell, G, Veesler, D. | | 登録日 | 2020-08-20 | | 公開日 | 2020-10-14 | | 最終更新日 | 2021-01-27 | | 実験手法 | ELECTRON MICROSCOPY (3.6 Å) | | 主引用文献 | Mapping Neutralizing and Immunodominant Sites on the SARS-CoV-2 Spike Receptor-Binding Domain by Structure-Guided High-Resolution Serology.
Cell, 183, 2020
|
|
7JXD
 
 | | Mapping neutralizing and immunodominant sites on the SARS-CoV-2 spike receptor-binding domain by structure-guided high-resolution serology | | 分子名称: | S2A4 antigen-binding (Fab) fragment | | 著者 | Park, Y.J, Tortorici, M.A, Walls, A.C, Czudnochowski, N, Seattle Structural Genomics Center for Infectious Disease (SSGCID), Snell, G, Veesler, D. | | 登録日 | 2020-08-27 | | 公開日 | 2020-10-14 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Mapping Neutralizing and Immunodominant Sites on the SARS-CoV-2 Spike Receptor-Binding Domain by Structure-Guided High-Resolution Serology.
Cell, 183, 2020
|
|
7JXE
 
 | | Mapping neutralizing and immunodominant sites on the SARS-CoV-2 spike receptor-binding domain by structure-guided high-resolution serology | | 分子名称: | S2X35 antigen-binding (Fab) fragment | | 著者 | Tortorici, M.A, Park, Y.J, Walls, A.C, Czudnochowski, N, Seattle Structural Genomics Center for Infectious Disease (SSGCID), Snell, G, Veesler, D. | | 登録日 | 2020-08-27 | | 公開日 | 2020-10-14 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.043 Å) | | 主引用文献 | Mapping Neutralizing and Immunodominant Sites on the SARS-CoV-2 Spike Receptor-Binding Domain by Structure-Guided High-Resolution Serology.
Cell, 183, 2020
|
|
