4PIM
 
 | |
6Q7U
 
 | |
6Q7W
 
 | |
6Q7V
 
 | |
4PIN
 
 | | Ergothioneine-biosynthetic methyltransferase EgtD in complex with N,N-dimethylhistidine | | 分子名称: | Histidine-specific methyltransferase EgtD, N,N-dimethyl-L-histidine, PHOSPHATE ION | | 著者 | Vit, A, Seebeck, F.P, Blankenfeldt, W. | | 登録日 | 2014-05-09 | | 公開日 | 2014-12-03 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Ergothioneine Biosynthetic Methyltransferase EgtD Reveals the Structural Basis of Aromatic Amino Acid Betaine Biosynthesis.
Chembiochem, 16, 2015
|
|
4PIO
 
 | | Ergothioneine-biosynthetic methyltransferase EgtD in complex with N,N-dimethylhistidine and SAH | | 分子名称: | CHLORIDE ION, Histidine-specific methyltransferase EgtD, MAGNESIUM ION, ... | | 著者 | Vit, A, Seebeck, F.P, Blankenfeldt, W. | | 登録日 | 2014-05-09 | | 公開日 | 2014-12-03 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.506 Å) | | 主引用文献 | Ergothioneine Biosynthetic Methyltransferase EgtD Reveals the Structural Basis of Aromatic Amino Acid Betaine Biosynthesis.
Chembiochem, 16, 2015
|
|
2Q0I
 
 | | Structure of Pseudomonas Quinolone Signal Response Protein PqsE | | 分子名称: | BENZOIC ACID, FE (III) ION, Quinolone signal response protein | | 著者 | Yu, S, Jensen, V, Feldmann, I, Haussler, S, Blankenfeldt, W. | | 登録日 | 2007-05-22 | | 公開日 | 2008-06-03 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (1.57 Å) | | 主引用文献 | Structure elucidation and preliminary assessment of hydrolase activity of PqsE, the Pseudomonas quinolone signal (PQS) response protein.
Biochemistry, 48, 2009
|
|
2Q0J
 
 | | Structure of Pseudomonas Quinolone Signal Response Protein PqsE | | 分子名称: | BENZOIC ACID, FE (III) ION, Quinolone signal response protein | | 著者 | Yu, S, Jensen, V, Feldmann, I, Haussler, S, Blankenfeldt, W. | | 登録日 | 2007-05-22 | | 公開日 | 2008-06-03 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Structure elucidation and preliminary assessment of hydrolase activity of PqsE, the Pseudomonas quinolone signal (PQS) response protein.
Biochemistry, 48, 2009
|
|
4ZFK
 
 | | Ergothioneine-biosynthetic Ntn hydrolase EgtC with glutamine | | 分子名称: | 1,2-ETHANEDIOL, Amidohydrolase EgtC, GLUTAMINE | | 著者 | Vit, A, Seebeck, F.P, Blankenfeldt, W. | | 登録日 | 2015-04-21 | | 公開日 | 2015-07-01 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.82 Å) | | 主引用文献 | Structure of the Ergothioneine-Biosynthesis Amidohydrolase EgtC.
Chembiochem, 16, 2015
|
|
4ZFJ
 
 | | Ergothioneine-biosynthetic Ntn hydrolase EgtC, apo form | | 分子名称: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 3,6,9,12,15,18,21,24,27-NONAOXANONACOSANE-1,29-DIOL, Amidohydrolase EgtC | | 著者 | Vit, A, Seebeck, F.P, Blankenfeldt, W. | | 登録日 | 2015-04-21 | | 公開日 | 2015-07-01 | | 最終更新日 | 2017-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Structure of the Ergothioneine-Biosynthesis Amidohydrolase EgtC.
Chembiochem, 16, 2015
|
|
4ZFL
 
 | |
1KCZ
 
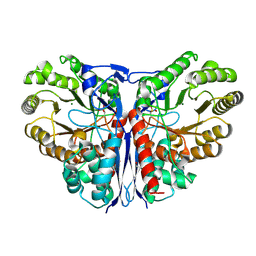 | | Crystal Structure of beta-methylaspartase from Clostridium tetanomorphum. Mg-complex. | | 分子名称: | 1,2-ETHANEDIOL, MAGNESIUM ION, beta-methylaspartase | | 著者 | Asuncion, M, Blankenfeldt, W, Barlow, J.N, Gani, D, Naismith, J.H. | | 登録日 | 2001-11-12 | | 公開日 | 2001-12-19 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | The structure of 3-methylaspartase from Clostridium tetanomorphum functions via the common enolase chemical step.
J.Biol.Chem., 277, 2002
|
|
1KD0
 
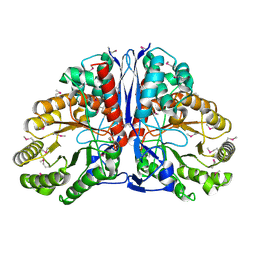 | | Crystal Structure of beta-methylaspartase from Clostridium tetanomorphum. Apo-structure. | | 分子名称: | 1,2-ETHANEDIOL, beta-methylaspartase | | 著者 | Asuncion, M, Blankenfeldt, W, Barlow, J.N, Gani, D, Naismith, J.H. | | 登録日 | 2001-11-12 | | 公開日 | 2001-12-19 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | The structure of 3-methylaspartase from Clostridium tetanomorphum functions via the common enolase chemical step.
J.Biol.Chem., 277, 2002
|
|
7QA0
 
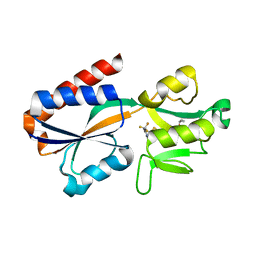 | |
7QAV
 
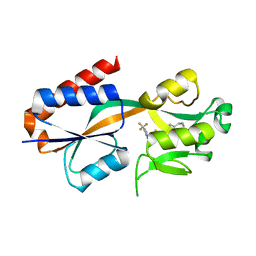 | |
7QA3
 
 | |
7QY3
 
 | | Crystal structure of the halohydrin dehalogenase HheG D114C mutant cross-linked with BMOE | | 分子名称: | 1,1'-ethane-1,2-diylbis(1H-pyrrole-2,5-dione), Putative oxidoreductase, SULFATE ION | | 著者 | Henke, S, Blankenfeldt, W, Schallmey, A. | | 登録日 | 2022-01-27 | | 公開日 | 2022-03-09 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2.72 Å) | | 主引用文献 | Biocatalytically active and stable cross-linked enzyme crystals of halohydrin dehalogenase HheG by protein engineering
Chemcatchem, 2022
|
|
7R3F
 
 | | Monomeric PqsE mutant E187R | | 分子名称: | 2-aminobenzoylacetyl-CoA thioesterase, BENZOIC ACID, CACODYLATE ION, ... | | 著者 | Borgert, S.R, Schmelz, S, Blankenfeldt, W. | | 登録日 | 2022-02-07 | | 公開日 | 2022-12-14 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | Moonlighting chaperone activity of the enzyme PqsE contributes to RhlR-controlled virulence of Pseudomonas aeruginosa.
Nat Commun, 13, 2022
|
|
7R3G
 
 | |
7R3I
 
 | |
7R3H
 
 | |
7R3J
 
 | | Nativ complex of PqsE and RhlR with the synthetic antagonist mBTL | | 分子名称: | 2-aminobenzoylacetyl-CoA thioesterase, 4-(3-bromophenoxy)-N-[(3S)-2-oxothiolan-3-yl]butanamide, FE (III) ION, ... | | 著者 | Borgert, S.R, Schmelz, S, Blankenfeldt, W. | | 登録日 | 2022-02-07 | | 公開日 | 2022-12-14 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (3.06 Å) | | 主引用文献 | Moonlighting chaperone activity of the enzyme PqsE contributes to RhlR-controlled virulence of Pseudomonas aeruginosa.
Nat Commun, 13, 2022
|
|
7R3E
 
 | | Fusion construct of PqsE and RhlR in complex with the synthetic antagonist mBTL | | 分子名称: | 2-aminobenzoylacetyl-CoA thioesterase,Regulatory protein RhlR, 4-(3-bromophenoxy)-N-[(3S)-2-oxothiolan-3-yl]butanamide, FE (III) ION | | 著者 | Borgert, S.R, Schmelz, S, Blankenfeldt, W. | | 登録日 | 2022-02-07 | | 公開日 | 2022-12-14 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (3.46 Å) | | 主引用文献 | Moonlighting chaperone activity of the enzyme PqsE contributes to RhlR-controlled virulence of Pseudomonas aeruginosa.
Nat Commun, 13, 2022
|
|
5JGL
 
 | | Crystal structure of GtmA in complex with S-Adenosylmethionine | | 分子名称: | S-ADENOSYLMETHIONINE, SODIUM ION, UbiE/COQ5 family methyltransferase, ... | | 著者 | Dolan, S.K, Bock, T, Hering, V, Jones, G.W, Blankenfeldt, W, Doyle, S. | | 登録日 | 2016-04-20 | | 公開日 | 2017-03-01 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.28 Å) | | 主引用文献 | Structural, mechanistic and functional insight into gliotoxinbis-thiomethylation inAspergillus fumigatus.
Open Biol, 7, 2017
|
|
3UI5
 
 | | Crystal structure of human Parvulin 14 | | 分子名称: | (4S,5S)-1,2-DITHIANE-4,5-DIOL, Peptidyl-prolyl cis-trans isomerase NIMA-interacting 4, SODIUM ION, ... | | 著者 | Mueller, J.W, Link, N.M, Matena, A, Hoppstock, L, Rueppel, A, Bayer, P, Blankenfeldt, W. | | 登録日 | 2011-11-04 | | 公開日 | 2011-12-07 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | Crystallographic proof for an extended hydrogen-bonding network in small prolyl isomerases.
J.Am.Chem.Soc., 133, 2011
|
|
