7XS8
 
 | |
7XSA
 
 | |
7XSC
 
 | |
7XSB
 
 | |
8Q68
 
 | | Crystal structure of TEAD1-YBD in complex with irreversible compound SWTX-143 | | 分子名称: | Transcriptional enhancer factor TEF-1, ~{N}-[(3~{S})-5-azanyl-1-[4-(trifluoromethyl)phenyl]-3,4-dihydro-2~{H}-quinolin-3-yl]propanamide | | 著者 | Ciesielski, F, Spieser, S.A.H, Marchand, A, Gwaltney, S.L. | | 登録日 | 2023-08-11 | | 公開日 | 2023-10-04 | | 最終更新日 | 2024-11-20 | | 実験手法 | X-RAY DIFFRACTION (1.58 Å) | | 主引用文献 | A Novel Irreversible TEAD Inhibitor, SWTX-143, Blocks Hippo Pathway Transcriptional Output and Causes Tumor Regression in Preclinical Mesothelioma Models.
Mol.Cancer Ther., 23, 2024
|
|
5JQG
 
 | | An apo tubulin-RB-TTL complex structure used for side-by-side comparison | | 分子名称: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, CHLORIDE ION, ... | | 著者 | Wang, Y.X, Naismith, J.H, Zhu, X. | | 登録日 | 2016-05-04 | | 公開日 | 2016-05-18 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.24 Å) | | 主引用文献 | Pironetin reacts covalently with cysteine-316 of alpha-tubulin to destabilize microtubule
Nat Commun, 7, 2016
|
|
1VK1
 
 | | Conserved hypothetical protein from Pyrococcus furiosus Pfu-392566-001 | | 分子名称: | Conserved hypothetical protein, DI(HYDROXYETHYL)ETHER, PHOSPHATE ION, ... | | 著者 | Shah, A, Liu, Z.J, Tempel, W, Chen, L, Lee, D, Yang, H, Chang, J, Zhao, M, Ng, J, Rose, J, Brereton, P.S, Izumi, M, Jenney Jr, F.E, Poole II, F.L, Shah, C, Sugar, F.J, Adams, M.W.W, Richardson, D.C, Richardson, J.S, Wang, B.C, Southeast Collaboratory for Structural Genomics (SECSG) | | 登録日 | 2004-04-13 | | 公開日 | 2004-08-10 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.2 Å) | | 主引用文献 | (NZ)CH...O contacts assist crystallization of a ParB-like nuclease.
Bmc Struct.Biol., 7, 2007
|
|
7FBJ
 
 | | Crystal structure of SARS-CoV-2 receptor binding domain in complex with neutralizing nanobody 17F6 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, New antigen receptor variable domain, ... | | 著者 | Zhu, J, Xu, T, Feng, B, Liu, J. | | 登録日 | 2021-07-11 | | 公開日 | 2022-07-13 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (2.85 Å) | | 主引用文献 | A Class of Shark-Derived Single-Domain Antibodies can Broadly Neutralize SARS-Related Coronaviruses and the Structural Basis of Neutralization and Omicron Escape.
Small Methods, 6, 2022
|
|
7FBK
 
 | | Crystal structure of SARS-CoV-2 receptor binding domain N501Y mutant in complex with neutralizing nanobody 20G6 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, New antigen receptor variable domain, Spike protein S1 | | 著者 | Zhu, J, Xu, T, Feng, B, Liu, J. | | 登録日 | 2021-07-11 | | 公開日 | 2022-07-13 | | 最終更新日 | 2024-11-06 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | A Class of Shark-Derived Single-Domain Antibodies can Broadly Neutralize SARS-Related Coronaviruses and the Structural Basis of Neutralization and Omicron Escape.
Small Methods, 6, 2022
|
|
4QR5
 
 | | Brd4 Bromodomain 1 complex with its novel inhibitors | | 分子名称: | Bromodomain-containing protein 4, N-[3-(cyclopentylsulfamoyl)-5-(2-oxo-2,3-dihydro-1,3-thiazol-4-yl)phenyl]cyclopropanecarboxamide | | 著者 | Xiong, B, Cao, D.Y, Chen, T.T, Xu, Y.C. | | 登録日 | 2014-06-30 | | 公開日 | 2015-07-01 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (1.41 Å) | | 主引用文献 | Fragment-based drug discovery of 2-thiazolidinones as BRD4 inhibitors: 2. Structure-based optimization
J.Med.Chem., 58, 2015
|
|
4QR3
 
 | | Brd4 Bromodomain 1 complex with its novel inhibitors | | 分子名称: | Bromodomain-containing protein 4, N-cyclopentyl-3-(2-oxo-2,3-dihydro-1,3-thiazol-4-yl)benzenesulfonamide | | 著者 | Xiong, B, Cao, D.Y, Chen, T.T, Xu, Y.C. | | 登録日 | 2014-06-30 | | 公開日 | 2015-07-01 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (1.374 Å) | | 主引用文献 | Fragment-based drug discovery of 2-thiazolidinones as BRD4 inhibitors: 2. Structure-based optimization
J.Med.Chem., 58, 2015
|
|
4QR4
 
 | | Brd4 Bromodomain 1 complex with its novel inhibitors | | 分子名称: | 2-chloro-N-cyclopentyl-5-(2-oxo-2,3-dihydro-1,3-thiazol-4-yl)benzenesulfonamide, Bromodomain-containing protein 4 | | 著者 | Xiong, B, Cao, D.Y, Chen, T.T, Xu, Y.C. | | 登録日 | 2014-06-30 | | 公開日 | 2015-07-01 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (1.28 Å) | | 主引用文献 | Fragment-based drug discovery of 2-thiazolidinones as BRD4 inhibitors: 2. Structure-based optimization
J.Med.Chem., 58, 2015
|
|
9JG6
 
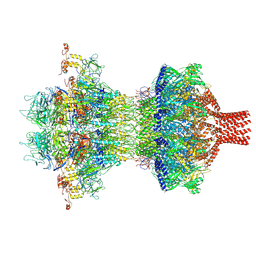 | | The tail-complex structure of phage P22 | | 分子名称: | Endorhamnosidase, P22 tail accessory factor, Phage stabilisation protein, ... | | 著者 | Liu, H.R, Xiao, H. | | 登録日 | 2024-09-06 | | 公開日 | 2025-02-26 | | 最終更新日 | 2025-04-30 | | 実験手法 | ELECTRON MICROSCOPY (3.21 Å) | | 主引用文献 | Structure of the scaffolding protein and portal within the bacteriophage P22 procapsid provides insights into the self-assembly process.
Plos Biol., 23, 2025
|
|
9KYV
 
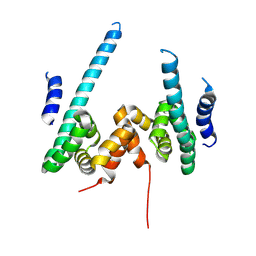 | | The scaffold dimer of phage P22 | | 分子名称: | Scaffolding protein | | 著者 | Liu, H.R, Xiao, H. | | 登録日 | 2024-12-09 | | 公開日 | 2025-02-26 | | 最終更新日 | 2025-04-30 | | 実験手法 | ELECTRON MICROSCOPY (4.9 Å) | | 主引用文献 | Structure of the scaffolding protein and portal within the bacteriophage P22 procapsid provides insights into the self-assembly process.
Plos Biol., 23, 2025
|
|
9KYW
 
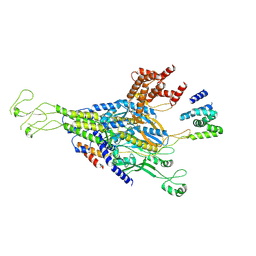 | | The scaffold C-loop of phage P22 | | 分子名称: | Portal protein, Scaffolding protein | | 著者 | Liu, H.R, Xiao, H. | | 登録日 | 2024-12-09 | | 公開日 | 2025-02-26 | | 最終更新日 | 2025-04-30 | | 実験手法 | ELECTRON MICROSCOPY (6.7 Å) | | 主引用文献 | Structure of the scaffolding protein and portal within the bacteriophage P22 procapsid provides insights into the self-assembly process.
Plos Biol., 23, 2025
|
|
9JGA
 
 | | P22 procapsid portal | | 分子名称: | Portal protein | | 著者 | Liu, H.R, Xiao, H. | | 登録日 | 2024-09-06 | | 公開日 | 2025-02-26 | | 最終更新日 | 2025-04-30 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | Structure of the scaffolding protein and portal within the bacteriophage P22 procapsid provides insights into the self-assembly process.
Plos Biol., 23, 2025
|
|
9KYY
 
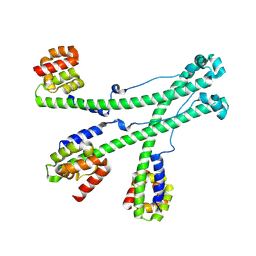 | | The scaffold trimer of phage P22 | | 分子名称: | Scaffolding protein | | 著者 | Liu, H.R, Xiao, H. | | 登録日 | 2024-12-09 | | 公開日 | 2025-02-26 | | 最終更新日 | 2025-04-30 | | 実験手法 | ELECTRON MICROSCOPY (6.9 Å) | | 主引用文献 | Structure of the scaffolding protein and portal within the bacteriophage P22 procapsid provides insights into the self-assembly process.
Plos Biol., 23, 2025
|
|
9KYX
 
 | | The scaffold tetramer of phage P22 | | 分子名称: | Scaffolding protein | | 著者 | Liu, H.R, Xiao, H. | | 登録日 | 2024-12-09 | | 公開日 | 2025-02-26 | | 最終更新日 | 2025-04-30 | | 実験手法 | ELECTRON MICROSCOPY (6.9 Å) | | 主引用文献 | Structure of the scaffolding protein and portal within the bacteriophage P22 procapsid provides insights into the self-assembly process.
Plos Biol., 23, 2025
|
|
1GHR
 
 | |
9MZA
 
 | | Chemically Hijacked BCL6-TCIP3-p300 Complex | | 分子名称: | 1-{1-[5-({1-[5-chloro-4-({8-methoxy-1-methyl-3-[2-(methylamino)-2-oxoethoxy]-2-oxo-1,2-dihydroquinolin-6-yl}amino)pyrimidin-2-yl]piperidine-4-carbonyl}amino)pentanoyl]piperidin-4-yl}-3-[(6M)-7-(difluoromethyl)-6-(1-methyl-1H-pyrazol-4-yl)-3,4-dihydroquinolin-1(2H)-yl]-N-methyl-1,4,6,7-tetrahydro-5H-pyrazolo[4,3-c]pyridine-5-carboxamide, B-cell lymphoma 6 protein, Histone acetyltransferase p300 | | 著者 | Hinshaw, S.M, Gray, N.S, Nix, M.N, Gourisankar, S, Martinez, M, Crabtree, G.R. | | 登録日 | 2025-01-22 | | 公開日 | 2025-04-16 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | A Bivalent Molecular Glue Linking Lysine Acetyltransferases to Oncogene-induced Cell Death.
Biorxiv, 2025
|
|
3UBU
 
 | | Crystal structure of agkisacucetin, a GpIb-binding snaclec (snake C-type lectin) that inhibits platelet | | 分子名称: | Agglucetin subunit alpha-1, Agglucetin subunit beta-2, GLYCEROL, ... | | 著者 | Gao, Y, Ge, H, Chen, H, Li, H, Liu, Y, Niu, L, Teng, M. | | 登録日 | 2011-10-25 | | 公開日 | 2012-04-11 | | 最終更新日 | 2025-03-26 | | 実験手法 | X-RAY DIFFRACTION (1.91 Å) | | 主引用文献 | Crystal structure of agkisacucetin, a Gpib-binding snake C-type lectin that inhibits platelet adhesion and aggregation.
Proteins, 2012
|
|
6LZH
 
 | | Crystal structure of Alpha/beta hydrolase GrgF from Penicillium sp. sh18 | | 分子名称: | GrgF, SODIUM ION | | 著者 | Wang, H, Yu, J, Wang, W.G, Matsuda, Y, Yao, M. | | 登録日 | 2020-02-19 | | 公開日 | 2020-06-24 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Molecular Basis for the Biosynthesis of an Unusual Chain-Fused Polyketide, Gregatin A.
J.Am.Chem.Soc., 142, 2020
|
|
4R8T
 
 | | Structure of JEV protease | | 分子名称: | CHLORIDE ION, NS3, Serine protease subunit NS2B | | 著者 | Nair, D.T, Weinert, T, Wang, M, Olieric, V. | | 登録日 | 2014-09-03 | | 公開日 | 2014-12-24 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.133 Å) | | 主引用文献 | Fast native-SAD phasing for routine macromolecular structure determination.
Nat.Methods, 12, 2015
|
|
2XPX
 
 | | Crystal structure of BHRF1:Bak BH3 complex | | 分子名称: | 1,2-ETHANEDIOL, APOPTOSIS REGULATOR BHRF1, BCL-2 HOMOLOGOUS ANTAGONIST/KILLER, ... | | 著者 | Kvansakul, M, Huang, D.C.S, Colman, P.M. | | 登録日 | 2010-08-31 | | 公開日 | 2011-01-26 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.05 Å) | | 主引用文献 | Structural Basis for Apoptosis Inhibition by Epstein-Barr Virus Bhrf1.
Plos Pathog., 6, 2010
|
|
2WH6
 
 | | Crystal structure of anti-apoptotic BHRF1 in complex with the Bim BH3 domain | | 分子名称: | BCL-2-LIKE PROTEIN 11, BROMIDE ION, EARLY ANTIGEN PROTEIN R, ... | | 著者 | Kvansakul, M, Huang, D.C.S, Colman, P.M. | | 登録日 | 2009-05-01 | | 公開日 | 2010-05-26 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Structural basis for apoptosis inhibition by Epstein-Barr virus BHRF1.
PLoS Pathog., 6, 2010
|
|
