4JGC
 
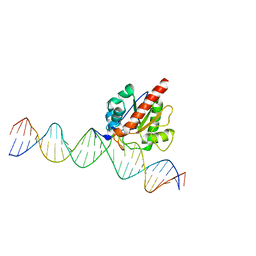 | | Human TDG N140A mutant IN A COMPLEX WITH 5-carboxylcytosine (5caC) | | 分子名称: | 4-amino-2-oxo-1,2-dihydropyrimidine-5-carboxylic acid, G/T mismatch-specific thymine DNA glycosylase, oligonucleotide, ... | | 著者 | Hashimoto, H, Zhang, X, Cheng, X. | | 登録日 | 2013-02-28 | | 公開日 | 2013-05-29 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.582 Å) | | 主引用文献 | Activity and crystal structure of human thymine DNA glycosylase mutant N140A with 5-carboxylcytosine DNA at low pH.
Dna Repair, 12, 2013
|
|
6NT9
 
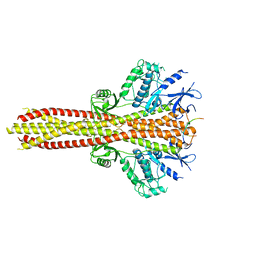 | | Cryo-EM structure of the complex between human TBK1 and chicken STING | | 分子名称: | Serine/threonine-protein kinase TBK1, Stimulator of interferon genes protein | | 著者 | Shang, G, Zhang, C, Chen, Z.J, Bai, X, Zhang, X. | | 登録日 | 2019-01-28 | | 公開日 | 2019-03-06 | | 最終更新日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | Structural basis of STING binding with and phosphorylation by TBK1.
Nature, 567, 2019
|
|
3QO2
 
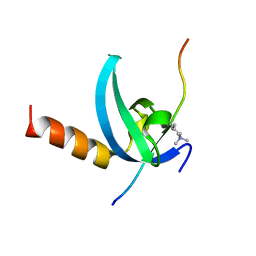 | | Structural insights for MPP8 chromodomain interaction with histone H3 lysine 9 | | 分子名称: | 1,2-ETHANEDIOL, Histone H3 peptide, M-phase phosphoprotein 8 | | 著者 | Chang, Y, Horton, J.R, Bedford, M.T, Zhang, X, Cheng, X. | | 登録日 | 2011-02-09 | | 公開日 | 2011-04-06 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.49 Å) | | 主引用文献 | Structural insights for MPP8 chromodomain interaction with histone H3 lysine 9: potential effect of phosphorylation on methyl-lysine binding.
J.Mol.Biol., 408, 2011
|
|
1S3S
 
 | | Crystal structure of AAA ATPase p97/VCP ND1 in complex with p47 C | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, Transitional endoplasmic reticulum ATPase (TER ATPase) (15S Mg(2+)- ATPase p97 subunit) (Valosin containing protein) (VCP) [Contains: Valosin], p47 protein | | 著者 | Dreveny, I, Kondo, H, Uchiyama, K, Shaw, A, Zhang, X, Freemont, P.S. | | 登録日 | 2004-01-14 | | 公開日 | 2004-03-30 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Structural basis of the interaction between the AAA ATPase p97/VCP and its adaptor protein p47.
Embo J., 23, 2004
|
|
2KA9
 
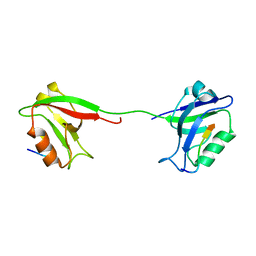 | | Solution structure of PSD-95 PDZ12 complexed with cypin peptide | | 分子名称: | Disks large homolog 4, cypin peptide | | 著者 | Wang, W.N, Weng, J.W, Zhang, X, Liu, M.L, Zhang, M.J. | | 登録日 | 2008-11-03 | | 公開日 | 2009-06-23 | | 最終更新日 | 2024-05-29 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Creating conformational entropy by increasing interdomain mobility in ligand binding regulation: a revisit to N-terminal tandem PDZ domains of PSD-95
J.Am.Chem.Soc., 131, 2009
|
|
3RC0
 
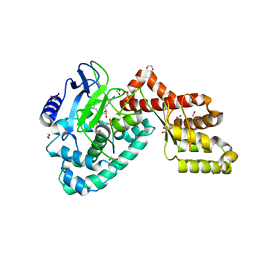 | | Human SETD6 in complex with RelA Lys310 peptide | | 分子名称: | 1,2-ETHANEDIOL, N-lysine methyltransferase SETD6, S-ADENOSYLMETHIONINE, ... | | 著者 | Chang, Y, Levy, D, Horton, J.R, Peng, J, Zhang, X, Gozani, O, Cheng, X. | | 登録日 | 2011-03-30 | | 公開日 | 2011-05-25 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.19 Å) | | 主引用文献 | Structural basis of SETD6-mediated regulation of the NF-kB network via methyl-lysine signaling.
Nucleic Acids Res., 39, 2011
|
|
4QOS
 
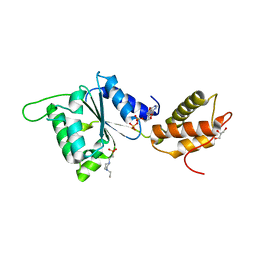 | | CRYSTAL STRUCTURE OF PSPF(1-265) E108Q MUTANT bound to ADP | | 分子名称: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ADENOSINE-5'-DIPHOSPHATE, GLYCEROL, ... | | 著者 | Darbari, V.C, Lawton, E, Lu, D, Burrows, P.C, Wiesler, S, Joly, N, Zhang, N, Zhang, X, Buck, M. | | 登録日 | 2014-06-20 | | 公開日 | 2014-08-06 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.42 Å) | | 主引用文献 | Molecular basis of nucleotide-dependent substrate engagement and remodeling by an AAA+ activator.
Nucleic Acids Res., 42, 2014
|
|
4QNM
 
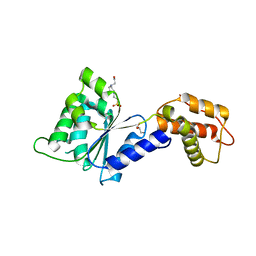 | | CRYSTAL STRUCTURE of PSPF(1-265) E108Q MUTANT | | 分子名称: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, GLYCEROL, Psp operon transcriptional activator | | 著者 | Darbari, V.C, Lawton, E, Lu, D, Burrows, P.C, Wiesler, S, Joly, N, Zhang, N, Zhang, X, Buck, M. | | 登録日 | 2014-06-18 | | 公開日 | 2014-08-06 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.628 Å) | | 主引用文献 | Molecular basis of nucleotide-dependent substrate engagement and remodeling by an AAA+ activator.
Nucleic Acids Res., 42, 2014
|
|
7XU2
 
 | | Structure of SARS-CoV-2 Spike Protein with Engineered x3 Disulfide (x3(D427C, V987C) and single Arg S1/S2 cleavage site), Locked-2 Conformation | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BILIVERDINE IX ALPHA, ... | | 著者 | Qu, K, Chen, Q, Ciazynska, K.A, Liu, B, Zhang, X, Wang, J, He, Y, Guan, J, He, J, Liu, T, Zhang, X, Carter, A.P, Xiong, X, Briggs, J.A.G. | | 登録日 | 2022-05-18 | | 公開日 | 2022-07-20 | | 最終更新日 | 2022-08-17 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Engineered disulfide reveals structural dynamics of locked SARS-CoV-2 spike.
Plos Pathog., 18, 2022
|
|
7XU0
 
 | | Structure of SARS-CoV-2 Spike Protein with Engineered x3 Disulfide (x3(D427C, V987C) and single Arg S1/S2 cleavage site), Locked-211 Conformation | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BILIVERDINE IX ALPHA, ... | | 著者 | Qu, K, Chen, Q, Ciazynska, K.A, Liu, B, Zhang, X, Wang, J, He, Y, Guan, J, He, J, Liu, T, Zhang, X, Carter, A.P, Xiong, X, Briggs, J.A.G. | | 登録日 | 2022-05-18 | | 公開日 | 2022-07-20 | | 最終更新日 | 2022-08-17 | | 実験手法 | ELECTRON MICROSCOPY (2.9 Å) | | 主引用文献 | Engineered disulfide reveals structural dynamics of locked SARS-CoV-2 spike.
Plos Pathog., 18, 2022
|
|
7XTZ
 
 | | Structure of SARS-CoV-2 Spike Protein with Engineered x3 Disulfide (x3(D427C, V987C) and single Arg S1/S2 cleavage site), Locked-1 Conformation | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BILIVERDINE IX ALPHA, ... | | 著者 | Qu, K, Chen, Q, Ciazynska, K.A, Liu, B, Zhang, X, Wang, J, He, Y, Guan, J, He, J, Liu, T, Zhang, X, Carter, A.P, Xiong, X, Briggs, J.A.G. | | 登録日 | 2022-05-18 | | 公開日 | 2022-07-20 | | 最終更新日 | 2022-08-17 | | 実験手法 | ELECTRON MICROSCOPY (2.8 Å) | | 主引用文献 | Engineered disulfide reveals structural dynamics of locked SARS-CoV-2 spike.
Plos Pathog., 18, 2022
|
|
7XU1
 
 | | Structure of SARS-CoV-2 Spike Protein with Engineered x3 Disulfide (x3(D427C, V987C) and single Arg S1/S2 cleavage site), Locked-122 Conformation | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BILIVERDINE IX ALPHA, ... | | 著者 | Qu, K, Chen, Q, Ciazynska, K.A, Liu, B, Zhang, X, Wang, J, He, Y, Guan, J, He, J, Liu, T, Zhang, X, Carter, A.P, Xiong, X, Briggs, J.A.G. | | 登録日 | 2022-05-18 | | 公開日 | 2022-07-20 | | 最終更新日 | 2022-08-17 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | Engineered disulfide reveals structural dynamics of locked SARS-CoV-2 spike.
Plos Pathog., 18, 2022
|
|
7XU4
 
 | | Structure of SARS-CoV-2 D614G Spike Protein with Engineered x3 Disulfide (x3(D427C, V987C) and single Arg S1/S2 cleavage site), Locked-2 Conformation | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BILIVERDINE IX ALPHA, ... | | 著者 | Qu, K, Chen, Q, Ciazynska, K.A, Liu, B, Zhang, X, Wang, J, He, Y, Guan, J, He, J, Liu, T, Zhang, X, Carter, A.P, Xiong, X, Briggs, J.A.G. | | 登録日 | 2022-05-18 | | 公開日 | 2022-07-20 | | 最終更新日 | 2022-08-17 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Engineered disulfide reveals structural dynamics of locked SARS-CoV-2 spike.
Plos Pathog., 18, 2022
|
|
7XU5
 
 | | Structure of SARS-CoV-2 D614G Spike Protein with Engineered x3 Disulfide (x3(D427C, V987C) and single Arg S1/S2 cleavage site), Closed Conformation | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BILIVERDINE IX ALPHA, ... | | 著者 | Qu, K, Chen, Q, Ciazynska, K.A, Liu, B, Zhang, X, Wang, J, He, Y, Guan, J, He, J, Liu, T, Zhang, X, Carter, A.P, Xiong, X, Briggs, J.A.G. | | 登録日 | 2022-05-18 | | 公開日 | 2022-07-20 | | 最終更新日 | 2022-08-17 | | 実験手法 | ELECTRON MICROSCOPY (3.1 Å) | | 主引用文献 | Engineered disulfide reveals structural dynamics of locked SARS-CoV-2 spike.
Plos Pathog., 18, 2022
|
|
7XU6
 
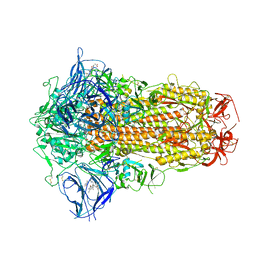 | | Structure of SARS-CoV-2 Spike Protein with Engineered x3 Disulfide (x3(D427C, V987C) and single Arg S1/S2 cleavage site), incubated in Low pH after 40-Day Storage in PBS, Locked-2 Conformation | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BILIVERDINE IX ALPHA, ... | | 著者 | Qu, K, Chen, Q, Ciazynska, K.A, Liu, B, Zhang, X, Wang, J, He, Y, Guan, J, He, J, Liu, T, Zhang, X, Carter, A.P, Xiong, X, Briggs, J.A.G. | | 登録日 | 2022-05-18 | | 公開日 | 2022-07-20 | | 最終更新日 | 2022-08-17 | | 実験手法 | ELECTRON MICROSCOPY (2.9 Å) | | 主引用文献 | Engineered disulfide reveals structural dynamics of locked SARS-CoV-2 spike.
Plos Pathog., 18, 2022
|
|
7XU3
 
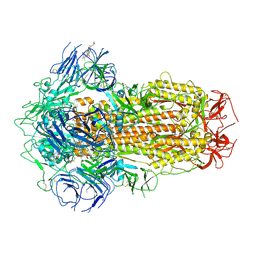 | | Structure of SARS-CoV-2 Spike Protein with Engineered x3 Disulfide (x3(D427C, V987C) and single Arg S1/S2 cleavage site), Closed Conformation | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BILIVERDINE IX ALPHA, ... | | 著者 | Qu, K, Chen, Q, Ciazynska, K.A, Liu, B, Zhang, X, Wang, J, He, Y, Guan, J, He, J, Liu, T, Zhang, X, Carter, A.P, Xiong, X, Briggs, J.A.G. | | 登録日 | 2022-05-18 | | 公開日 | 2022-07-20 | | 最終更新日 | 2022-08-17 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | Engineered disulfide reveals structural dynamics of locked SARS-CoV-2 spike.
Plos Pathog., 18, 2022
|
|
5CB3
 
 | | Structural Insights into the Mechanism of Escherichia coli Ymdb | | 分子名称: | ADENOSINE-5-DIPHOSPHORIBOSE, O-acetyl-ADP-ribose deacetylase | | 著者 | Zhang, W, Wang, C, Song, Y, Shao, C, Zhang, X, Zang, J. | | 登録日 | 2015-06-30 | | 公開日 | 2015-11-04 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Structural insights into the mechanism of Escherichia coli YmdB: A 2'-O-acetyl-ADP-ribose deacetylase
J.Struct.Biol., 192, 2015
|
|
5COA
 
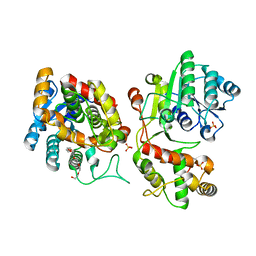 | | Crystal structure of iridoid synthase at 2.2-angstrom resolution | | 分子名称: | HEXAETHYLENE GLYCOL, Iridoid synthase, SULFATE ION | | 著者 | Qin, L, Zhu, Y, Ding, Z, Zhang, X, Ye, S, Zhang, R. | | 登録日 | 2015-07-20 | | 公開日 | 2016-03-09 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Structure of iridoid synthase in complex with NADP(+)/8-oxogeranial reveals the structural basis of its substrate specificity.
J.Struct.Biol., 194, 2016
|
|
7VVS
 
 | |
8EW2
 
 | |
8F49
 
 | |
8EW0
 
 | |
5COB
 
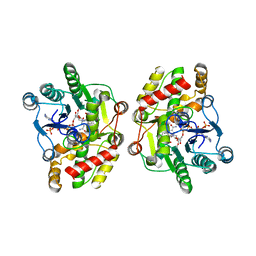 | | Crystal structure of iridoid synthase in complex with NADP+ and 8-oxogeranial at 2.65-angstrom resolution | | 分子名称: | (2E,6E)-2,6-dimethylocta-2,6-dienedial, Iridoid synthase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | 著者 | Qin, L, Zhu, Y, Ding, Z, Zhang, X, Ye, S, Zhang, R. | | 登録日 | 2015-07-20 | | 公開日 | 2016-03-09 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.65 Å) | | 主引用文献 | Structure of iridoid synthase in complex with NADP(+)/8-oxogeranial reveals the structural basis of its substrate specificity.
J.Struct.Biol., 194, 2016
|
|
5CB5
 
 | | Structural Insights into the Mechanism of Escherichia coli Ymdb | | 分子名称: | ACETATE ION, ADENOSINE-5-DIPHOSPHORIBOSE, O-acetyl-ADP-ribose deacetylase, ... | | 著者 | Zhang, W, Wang, C, Song, Y, Shao, C, Zhang, X, Zang, J. | | 登録日 | 2015-06-30 | | 公開日 | 2015-11-04 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Structural insights into the mechanism of Escherichia coli YmdB: A 2'-O-acetyl-ADP-ribose deacetylase
J.Struct.Biol., 192, 2015
|
|
5CMS
 
 | | Structural Insights into the Mechanism of Escherichia coli Ymdb | | 分子名称: | ADENOSINE-5-DIPHOSPHORIBOSE, O-acetyl-ADP-ribose deacetylase, SULFATE ION | | 著者 | Zhang, W, Wang, C, Song, Y, Shao, C, Zhang, X, Zang, J. | | 登録日 | 2015-07-17 | | 公開日 | 2015-11-04 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.98 Å) | | 主引用文献 | Structural insights into the mechanism of Escherichia coli YmdB: A 2'-O-acetyl-ADP-ribose deacetylase
J.Struct.Biol., 192, 2015
|
|
