3GRC
 
 | |
3DD5
 
 | | Glomerella cingulata E600-cutinase complex | | 分子名称: | Cutinase, DIETHYL PHOSPHONATE | | 著者 | Nyon, M.P, Rice, D.W, Berrisford, J.M, Hounslow, A.M, Moir, A.J.G, Huang, H, Nathan, S, Mahadi, N.M, Farah Diba, A.B, Craven, C.J. | | 登録日 | 2008-06-05 | | 公開日 | 2008-11-18 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Catalysis by Glomerella cingulata Cutinase Requires Conformational Cycling between the Active and Inactive States of Its Catalytic Triad
J.Mol.Biol., 385, 2009
|
|
3DCN
 
 | | Glomerella cingulata apo cutinase | | 分子名称: | Cutinase | | 著者 | Nyon, M.P, Rice, D.W, Berrisford, J.M, Hounslow, A.M, Moir, A.J.G, Huang, H, Nathan, S, Mahadi, N.M, Farah Diba, A.B, Craven, C.J. | | 登録日 | 2008-06-04 | | 公開日 | 2008-11-18 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Catalysis by Glomerella cingulata Cutinase Requires Conformational Cycling between the Active and Inactive States of Its Catalytic Triad
J.Mol.Biol., 385, 2009
|
|
3DEA
 
 | | Glomerella cingulata PETFP-cutinase complex | | 分子名称: | 1,1,1-trifluoro-3-[(2-phenylethyl)sulfanyl]propan-2-one, Cutinase | | 著者 | Nyon, M.P, Rice, D.W, Berrisford, J.M, Hounslow, A.M, Moir, A.J.G, Huang, H, Nathan, S, Mahadi, N.M, Farah Diba, A.B, Craven, C.J. | | 登録日 | 2008-06-09 | | 公開日 | 2008-11-18 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Catalysis by Glomerella cingulata Cutinase Requires Conformational Cycling between the Active and Inactive States of Its Catalytic Triad
J.Mol.Biol., 385, 2009
|
|
3GXV
 
 | | Three-dimensional structure of N-terminal domain of DnaB Helicase from Helicobacter pylori and its interactions with primase | | 分子名称: | Replicative DNA helicase | | 著者 | Kashav, T, Nitharwal, R, Syed, A.A, Gabdoulkhakov, A, Saenger, W, Dhar, K.S, Gourinath, S. | | 登録日 | 2009-04-03 | | 公開日 | 2010-01-26 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Three-dimensional structure of N-terminal domain of DnaB helicase and helicase-primase interactions in Helicobacter pylori
Plos One, 4, 2009
|
|
8GOD
 
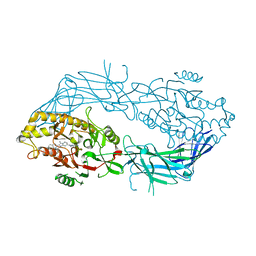 | | Co-crystal structure of Human Protein-arginine deiminase type-4 (PAD4) with small molecule inhibitor JBI-589 | | 分子名称: | Protein-arginine deiminase type-4, [(3~{R})-3-azanylpiperidin-1-yl]-[2-[1-[(4-fluorophenyl)methyl]indol-2-yl]-3-methyl-imidazo[1,2-a]pyridin-7-yl]methanone | | 著者 | Swaminathan, S, Birudukota, S, Vaithilingam, K, Kandan, S, Asaithambi, K, Kathiresan, N, Gosu, R, Rajagopal, S, Sadhu, N. | | 登録日 | 2022-08-24 | | 公開日 | 2023-03-29 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.88 Å) | | 主引用文献 | Alleviation of arthritis through prevention of neutrophil extracellular traps by an orally available inhibitor of protein arginine deiminase 4.
Sci Rep, 13, 2023
|
|
7TLT
 
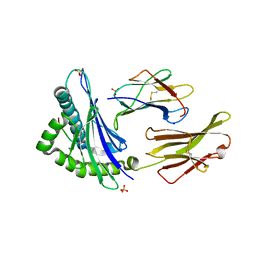 | | SARS-CoV-2 Spike-derived peptide S489-497 (YFPLQSYGF) presented by HLA-A*29:02 | | 分子名称: | Beta-2-microglobulin, HLA class I histocompatibility antigen, A alpha chain, ... | | 著者 | Murdolo, L.D, Szeto, C, Gras, S. | | 登録日 | 2022-01-18 | | 公開日 | 2022-10-12 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Ablation of CD8 + T cell recognition of an immunodominant epitope in SARS-CoV-2 Omicron variants BA.1, BA.2 and BA.3.
Nat Commun, 13, 2022
|
|
6B67
 
 | |
4DS7
 
 | |
6D45
 
 | | L89S Mutant of FeBMb Sperm Whale Myoglobin | | 分子名称: | Myoglobin, PROTOPORPHYRIN IX CONTAINING FE | | 著者 | Bhagi-Damodaran, A, Mirts, E.N, Sandoval, B, Lu, Y. | | 登録日 | 2018-04-17 | | 公開日 | 2018-05-23 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.779 Å) | | 主引用文献 | Heme redox potentials hold the key to reactivity differences between nitric oxide reductase and heme-copper oxidase.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
8RVE
 
 | | Vimentin intermediate filament | | 分子名称: | Vimentin | | 著者 | Eibauer, M, Medalia, O. | | 登録日 | 2024-02-01 | | 公開日 | 2024-04-10 | | 最終更新日 | 2024-07-03 | | 実験手法 | ELECTRON MICROSCOPY (7.2 Å) | | 主引用文献 | Vimentin filaments integrate low-complexity domains in a complex helical structure.
Nat.Struct.Mol.Biol., 31, 2024
|
|
7PQV
 
 | | MEK1 IN COMPLEX WITH COMPOUND 7 | | 分子名称: | 8-(2-chloranyl-4-methoxy-phenyl)-7-fluoranyl-1-piperidin-4-yl-imidazo[4,5-c]quinoline, CALCIUM ION, Dual specificity mitogen-activated protein kinase kinase 1, ... | | 著者 | Moebitz, H. | | 登録日 | 2021-09-20 | | 公開日 | 2022-03-16 | | 最終更新日 | 2024-06-19 | | 実験手法 | X-RAY DIFFRACTION (2.13 Å) | | 主引用文献 | Discovery of MAP855, an Efficacious and Selective MEK1/2 Inhibitor with an ATP-Competitive Mode of Action.
J.Med.Chem., 65, 2022
|
|
4TUN
 
 | |
1PVD
 
 | |
8EUA
 
 | | Structure of SARS-CoV2 PLpro bound to a covalent inhibitor | | 分子名称: | Papain-like protease nsp3, SULFATE ION, ZINC ION, ... | | 著者 | Mathews, I.I, Pokhrel, S, Wakatsuki, S. | | 登録日 | 2022-10-18 | | 公開日 | 2023-04-05 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (3.1 Å) | | 主引用文献 | Potent and selective covalent inhibition of the papain-like protease from SARS-CoV-2.
Nat Commun, 14, 2023
|
|
1PYD
 
 | |
8BGF
 
 | |
8DL9
 
 | |
8DLB
 
 | | Room temperature X-ray structure of SARS-CoV-2 main protease in complex with compound Z2799209083 | | 分子名称: | 1-[(5S)-5-(3,4-dimethoxyphenyl)-3-phenyl-4,5-dihydro-1H-pyrazol-1-yl]ethan-1-one, 3C-like proteinase | | 著者 | Kovalevsky, A.Y, Coates, L, Kneller, D.W. | | 登録日 | 2022-07-07 | | 公開日 | 2023-05-17 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | AI-Accelerated Design of Targeted Covalent Inhibitors for SARS-CoV-2.
J.Chem.Inf.Model., 63, 2023
|
|
8DMD
 
 | |
1BJR
 
 | | COMPLEX FORMED BETWEEN PROTEOLYTICALLY GENERATED LACTOFERRIN FRAGMENT AND PROTEINASE K | | 分子名称: | CALCIUM ION, LACTOFERRIN, PROTEINASE K | | 著者 | Singh, T.P, Sharma, S, Karthikeyan, S, Betzel, C, Bhatia, K.L. | | 登録日 | 1998-06-27 | | 公開日 | 1998-11-04 | | 最終更新日 | 2023-08-02 | | 実験手法 | X-RAY DIFFRACTION (2.44 Å) | | 主引用文献 | Crystal structure of a complex formed between proteolytically-generated lactoferrin fragment and proteinase K.
Proteins, 33, 1998
|
|
6V2F
 
 | | Crystal structure of the HIV capsid hexamer bound to the small molecule long-acting inhibitor, GS-6207 | | 分子名称: | HIV-1 capsid, N-[(1S)-1-(3-{4-chloro-3-[(methylsulfonyl)amino]-1-(2,2,2-trifluoroethyl)-1H-indazol-7-yl}-6-[3-methyl-3-(methylsulfonyl)but-1-yn-1-yl]pyridin-2-yl)-2-(3,5-difluorophenyl)ethyl]-2-[(3bS,4aR)-5,5-difluoro-3-(trifluoromethyl)-3b,4,4a,5-tetrahydro-1H-cyclopropa[3,4]cyclopenta[1,2-c]pyrazol-1-yl]acetamide | | 著者 | Appleby, T.C, Link, J.O, Yant, S.R, Villasenor, A.G, Somoza, J.R, Hu, E.Y, Schroeder, S.D, Cihlar, T. | | 登録日 | 2019-11-22 | | 公開日 | 2020-07-01 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Clinical targeting of HIV capsid protein with a long-acting small molecule.
Nature, 584, 2020
|
|
7SI9
 
 | | Room temperature X-ray structure of SARS-CoV-2 main protease (Mpro) in complex with PF-07321332 | | 分子名称: | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase | | 著者 | Kovalevsky, A, Kneller, D.W, Coates, L. | | 登録日 | 2021-10-12 | | 公開日 | 2021-10-20 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Covalent narlaprevir- and boceprevir-derived hybrid inhibitors of SARS-CoV-2 main protease
Nat Commun, 13, 2022
|
|
7TDU
 
 | | Joint X-ray/neutron structure of SARS-CoV-2 main protease (3CL Mpro) in complex with BBH-1 | | 分子名称: | (1R,2S,5S)-N-{(1S,2S)-1-(1,3-benzothiazol-2-yl)-1-hydroxy-3-[(3S)-2-oxo(1-~2~H)pyrrolidin-3-yl]propan-2-yl}-3-{N-[tert-butyl(~2~H)carbamoyl]-3-methyl-L-(N-~2~H)valyl}-6,6-dimethyl-3-azabicyclo[3.1.0]hexane-2-(~2~H)carboxamide, 3C-like proteinase | | 著者 | Kovalevsky, A, Kneller, D.W, Coates, L. | | 登録日 | 2022-01-03 | | 公開日 | 2022-03-02 | | 最終更新日 | 2024-04-03 | | 実験手法 | NEUTRON DIFFRACTION (1.85 Å), X-RAY DIFFRACTION | | 主引用文献 | Covalent narlaprevir- and boceprevir-derived hybrid inhibitors of SARS-CoV-2 main protease
Nat Commun, 13, 2022
|
|
6VND
 
 | | Quaternary Complex of human dihydroorotate dehydrogenase (DHODH) with flavin mononucleotide (FMN), orotic acid and AG-636 | | 分子名称: | 1-methyl-5-(2'-methyl[1,1'-biphenyl]-4-yl)-1H-benzotriazole-7-carboxylic acid, ACETATE ION, CHLORIDE ION, ... | | 著者 | Padyana, A, Jin, L. | | 登録日 | 2020-01-29 | | 公開日 | 2020-11-04 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.97 Å) | | 主引用文献 | Selective Vulnerability to Pyrimidine Starvation in Hematologic Malignancies Revealed by AG-636, a Novel Clinical-Stage Inhibitor of Dihydroorotate Dehydrogenase.
Mol.Cancer Ther., 19, 2020
|
|
