2FBL
 
 | | The crystal structure of the hypothetical protein NE1496 | | 分子名称: | SODIUM ION, hypothetical protein NE1496 | | 著者 | Lunin, V.V, Skarina, T, Onopriyenko, O, Binkowski, T.A, Joachimiak, A, Edwards, A.M, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2005-12-09 | | 公開日 | 2005-12-20 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | The crystal structure of the hypothetical protein NE1496
To be Published
|
|
2QQY
 
 | | Crystal structure of ferritin like, diiron-carboxylate proteins from Bacillus anthracis str. Ames | | 分子名称: | Sigma B operon | | 著者 | Kim, Y, Joachimiak, G, Wu, R, Patterson, S, Gornicki, P, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2007-07-27 | | 公開日 | 2007-08-14 | | 最終更新日 | 2017-10-25 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Crystal Structure of Ferritin like, Diiron-carboxylate Proteins from Bacillus anthracis str. Ames.
To be Published
|
|
2FBI
 
 | | The crystal structure of transcriptional regulator PA4135 | | 分子名称: | probable transcriptional regulator | | 著者 | Lunin, V.V, Evdokimova, E, Kudritska, M, Cuff, M.E, Joachimiak, A, Edwards, A.M, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2005-12-09 | | 公開日 | 2005-12-20 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | The crystal structure of transcriptional regulator PA4135
To be Published
|
|
2FBH
 
 | | The crystal structure of transcriptional regulator PA3341 | | 分子名称: | MERCURY (II) ION, SULFATE ION, ZINC ION, ... | | 著者 | Lunin, V.V, Evdokimova, E, Kudritska, M, Osipiuk, J, Joachimiak, A, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2005-12-09 | | 公開日 | 2005-12-20 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | The crystal structure of transcriptional regulator PA3341
To be Published
|
|
4ZQN
 
 | | Crystal Structure of the Catalytic Domain of the Inosine Monophosphate Dehydrogenase from Mycobacterium tuberculosis in the complex with IMP and the inhibitor P41 | | 分子名称: | 2-chloro-N,N-dimethyl-5-[({2-[3-(prop-1-en-2-yl)phenyl]propan-2-yl}carbamoyl)amino]benzamide, INOSINIC ACID, Inosine-5'-monophosphate dehydrogenase,Inosine-5'-monophosphate dehydrogenase | | 著者 | Kim, Y, Makowska-Grzyska, M, Gu, M, Kavitha, M, Hedstrom, L, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2015-05-10 | | 公開日 | 2015-06-17 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Mycobacterium tuberculosis IMPDH in Complexes with Substrates, Products and Antitubercular Compounds.
Plos One, 10, 2015
|
|
1HRU
 
 | | THE STRUCTURE OF THE YRDC GENE PRODUCT FROM E.COLI | | 分子名称: | PHOSPHATE ION, YRDC GENE PRODUCT | | 著者 | Teplova, M, Tereshko, V, Sanishvili, R, Joachimiak, A, Bushueva, T, Anderson, W.F, Egli, M, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2000-12-21 | | 公開日 | 2001-01-31 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | The structure of the yrdC gene product from Escherichia coli reveals a new fold and suggests a role in RNA binding.
Protein Sci., 9, 2000
|
|
4ZDN
 
 | | Streptomyces platensis isomigrastatin ketosynthase domain MgsF KS4 | | 分子名称: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, AT-less polyketide synthase, CHLORIDE ION | | 著者 | Chang, C, Li, H, Endres, M, Bingman, C.A, Yennamalli, R, Lohman, J.R, Ma, M, Shen, B, Phillips Jr, G.N, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | 登録日 | 2015-04-17 | | 公開日 | 2015-05-13 | | 最終更新日 | 2023-03-22 | | 実験手法 | X-RAY DIFFRACTION (2.509 Å) | | 主引用文献 | Structural and evolutionary relationships of "AT-less" type I polyketide synthase ketosynthases.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
1I3W
 
 | | ACTINOMYCIN D BINDING TO CGATCGATCG | | 分子名称: | 5'-D(*C*GP*AP*TP*CP*GP*AP*(BRU)P*CP*GP)-3', ACTINOMYCIN D | | 著者 | Robinson, H, Gao, Y.-G, Yang, X.-L, Sanishvili, R, Joachimiak, A, Wang, A.H.-J. | | 登録日 | 2001-02-17 | | 公開日 | 2001-05-21 | | 最終更新日 | 2024-07-10 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Crystallographic Analysis of a Novel Complex of Actinomycin D Bound to the DNA Decamer Cgatcgatcg.
Biochemistry, 40, 2001
|
|
4ZQM
 
 | | Crystal Structure of the Catalytic Domain of the Inosine Monophosphate Dehydrogenase from Mycobacterium tuberculosis in the complex with XMP and NAD | | 分子名称: | Inosine-5'-monophosphate dehydrogenase,Inosine-5'-monophosphate dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, XANTHOSINE-5'-MONOPHOSPHATE | | 著者 | Kim, Y, Maltseva, N, Makowska-Grzyska, M, Gu, M, Kavitha, M, Hedstrom, L, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2015-05-10 | | 公開日 | 2015-06-17 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (1.602 Å) | | 主引用文献 | Mycobacterium tuberculosis IMPDH in Complexes with Substrates, Products and Antitubercular Compounds.
Plos One, 10, 2015
|
|
4Z5P
 
 | | Crystal structure of the LnmA cytochrome P450 hydroxylase from the leinamycin biosynthetic pathway of Streptomyces atroolivaceus S-140 at 1.9 A resolution | | 分子名称: | Cytochrome P450 hydroxylase, PROTOPORPHYRIN IX CONTAINING FE, TRIETHYLENE GLYCOL | | 著者 | Ma, M, Lohman, J, Rudolf, J, Miller, M.D, Cao, H, Osipiuk, J, Babnigg, G, Phillips Jr, G.N, Joachimiak, A, Shen, B, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | 登録日 | 2015-04-02 | | 公開日 | 2015-07-29 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Crystal structure of the LnmA cytochrome P450 hydroxylase from the leinamycin biosynthetic pathway of Streptomyces atroolivaceus S-140
To be Published
|
|
4ZO4
 
 | | Dephospho-CoA kinase from Campylobacter jejuni. | | 分子名称: | BETA-MERCAPTOETHANOL, Dephospho-CoA kinase | | 著者 | Osipiuk, J, Zhou, M, Stam, J, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2015-05-06 | | 公開日 | 2015-05-13 | | 最終更新日 | 2019-12-11 | | 実験手法 | X-RAY DIFFRACTION (2.57 Å) | | 主引用文献 | Dephospho-CoA kinase from Campylobacter jejuni.
to be published
|
|
4ZPJ
 
 | | ABC transporter substrate-binding protein from Sphaerobacter thermophilus | | 分子名称: | CHLORIDE ION, Extracellular ligand-binding receptor, ZINC ION | | 著者 | OSIPIUK, J, Holowicki, J, Clancy, S, JOACHIMIAK, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2015-05-07 | | 公開日 | 2015-05-20 | | 最終更新日 | 2019-12-25 | | 実験手法 | X-RAY DIFFRACTION (2.24 Å) | | 主引用文献 | ABC transporter substrate-binding protein from Sphaerobacter thermophilus.
to be published
|
|
4ZQP
 
 | | Crystal Structure of the Catalytic Domain of the Inosine Monophosphate Dehydrogenase from Mycobacterium tuberculosis in the complex with IMP and the inhibitor MAD1 | | 分子名称: | 5'-O-({1-[(2E)-4-(4-hydroxy-6-methoxy-7-methyl-3-oxo-1,3-dihydro-2-benzofuran-5-yl)-2-methylbut-2-en-1-yl]-1H-1,2,3-triazol-4-yl}methyl)adenosine, GLYCEROL, INOSINIC ACID, ... | | 著者 | Kim, Y, Makowska-Grzyska, M, Gu, M, Kavitha, M, Hedstrom, L, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2015-05-10 | | 公開日 | 2015-06-17 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Mycobacterium tuberculosis IMPDH in Complexes with Substrates, Products and Antitubercular Compounds.
Plos One, 10, 2015
|
|
4ZR7
 
 | | The structure of a domain of a functionally unknown protein from Bacillus subtilis subsp. subtilis str. 168 | | 分子名称: | ACETATE ION, CHLORIDE ION, Sensor histidine kinase ResE | | 著者 | Tan, K, Li, H, Jedrzejczak, R, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2015-05-11 | | 公開日 | 2015-05-27 | | 最終更新日 | 2019-12-25 | | 実験手法 | X-RAY DIFFRACTION (1.86 Å) | | 主引用文献 | The structure of a domain of a functionally unknown protein from Bacillus subtilis subsp. subtilis str. 168
To Be Published
|
|
4ZNM
 
 | | Crystal structure of SgcC5 protein from Streptomyces globisporus (apo form) | | 分子名称: | C-domain type II peptide synthetase, CHLORIDE ION, SODIUM ION | | 著者 | Michalska, K, Bigelow, L, Jedrzejczak, R, Babnigg, G, Lohman, J, Ma, M, Rudolf, J, Chang, C.-Y, Shen, B, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | 登録日 | 2015-05-04 | | 公開日 | 2015-05-27 | | 最終更新日 | 2019-12-25 | | 実験手法 | X-RAY DIFFRACTION (1.998 Å) | | 主引用文献 | Crystal structure of SgcC5 protein from Streptomyces globisporus (apo form)
To Be Published
|
|
4ZQR
 
 | | Crystal Structure of the Catalytic Domain of the Inosine Monophosphate Dehydrogenase from Mycobacterium tuberculosis | | 分子名称: | GLYCEROL, Inosine-5'-monophosphate dehydrogenase,Inosine-5'-monophosphate dehydrogenase, PHOSPHATE ION, ... | | 著者 | Kim, Y, Makowska-Grzyska, M, Gu, M, Kavitha, M, Hedstrom, L, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2015-05-11 | | 公開日 | 2015-06-17 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (1.692 Å) | | 主引用文献 | Mycobacterium tuberculosis IMPDH in Complexes with Substrates, Products and Antitubercular Compounds.
Plos One, 10, 2015
|
|
4ZV9
 
 | | 2.00 Angstrom resolution crystal structure of an uncharacterized protein from Escherichia coli O157:H7 str. Sakai | | 分子名称: | DI(HYDROXYETHYL)ETHER, GLYCEROL, PHOSPHATE ION, ... | | 著者 | Halavaty, A.S, Wawrzak, Z, Filippova, E.V, Kiryukhina, O, Endres, M, Joachimiak, A, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2015-05-18 | | 公開日 | 2015-06-17 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | 2.00 Angstrom resolution crystal structure of an uncharacterized protein from Escherichia coli O157:H7 str. Sakai
To Be Published
|
|
5BMO
 
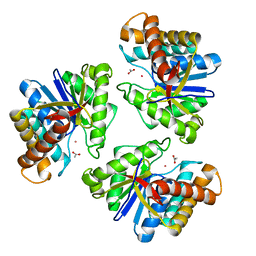 | | LnmX protein, a putative GlcNAc-PI de-N-acetylase from Streptomyces atroolivaceus | | 分子名称: | ACETATE ION, POTASSIUM ION, Putative uncharacterized protein LnmX | | 著者 | Osipiuk, J, Hatzos-Skintges, C, Cuff, M, Endres, M, Babnigg, G, Lohman, J, Ma, M, Rudolf, J, Chang, C.-Y, Shen, B, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | 登録日 | 2015-05-22 | | 公開日 | 2015-06-10 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.92 Å) | | 主引用文献 | LnmX protein, a putative GlcNAc-PI de-N-acetylase from Streptomyces atroolivaceus.
to be published
|
|
1YLQ
 
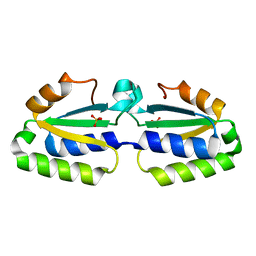 | | Crystal structure of putative nucleotidyltransferase | | 分子名称: | SULFATE ION, putative nucleotidyltransferase, hypothetical protein AF0614 | | 著者 | Chang, C, Joachimiak, A, Skarina, T, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2005-01-19 | | 公開日 | 2005-03-01 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2.016 Å) | | 主引用文献 | Crystal structure of Hypothetical protein AF0614, putative nucleotidyltransferase
To be Published
|
|
5C4Y
 
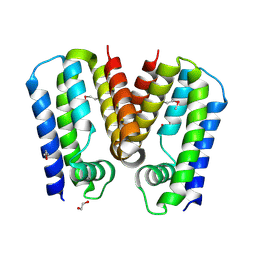 | | Crystal structure of putative TetR family transcription factor from Listeria monocytogenes | | 分子名称: | 1,2-ETHANEDIOL, Putative transcription regulator Lmo0852 | | 著者 | Chang, C, Tesar, C, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2015-06-18 | | 公開日 | 2015-07-08 | | 実験手法 | X-RAY DIFFRACTION (1.77 Å) | | 主引用文献 | Crystal structure of putative TetR family transcription factor from Listeria monocytogenes
to be published
|
|
1ZKE
 
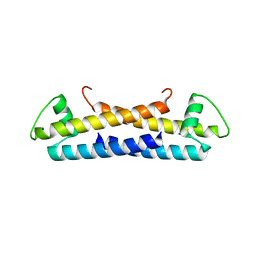 | | 1.6 A Crystal Structure of a Protein HP1531 of Unknown Function from Helicobacter pylori | | 分子名称: | Hypothetical protein HP1531, MAGNESIUM ION | | 著者 | Zhang, R, Skarina, T, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2005-05-02 | | 公開日 | 2005-06-28 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | 1.6A crystal structure of a hypothetical protein HP1531 from Helicobacter pylori 26695
To be Published
|
|
4ZQO
 
 | | Crystal Structure of the Catalytic Domain of the Inosine Monophosphate Dehydrogenase from Mycobacterium tuberculosis in the complex with IMP and the inhibitor Q67 | | 分子名称: | GLYCEROL, INOSINIC ACID, Inosine-5'-monophosphate dehydrogenase,Inosine-5'-monophosphate dehydrogenase, ... | | 著者 | Kim, Y, Makowska-Grzyska, M, Gu, M, Kavitha, M, Hedstrom, L, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2015-05-10 | | 公開日 | 2015-06-17 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (1.76 Å) | | 主引用文献 | Mycobacterium tuberculosis IMPDH in Complexes with Substrates, Products and Antitubercular Compounds.
Plos One, 10, 2015
|
|
4Z5Q
 
 | | Crystal structure of the LnmZ cytochrome P450 hydroxylase from the leinamycin biosynthetic pathway of Streptomyces atroolivaceus S-140 at 1.8 A resolution | | 分子名称: | 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, CHLORIDE ION, Cytochrome P450 hydroxylase, ... | | 著者 | Ma, M, Lohman, J, Rudolf, J, Miller, M.D, Cao, H, Osipiuk, J, Joachimiak, A, Phillips Jr, G.N, Shen, B, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | 登録日 | 2015-04-02 | | 公開日 | 2015-08-12 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.801 Å) | | 主引用文献 | Crystal structure of the LnmZ cytochrome P450 hydroxylase from the leinamycin biosynthetic pathway of Streptomyces atroolivaceus S-140
To be Published
|
|
5CNP
 
 | | X-ray crystal structure of Spermidine n1-acetyltransferase from Vibrio cholerae. | | 分子名称: | ISOPROPYL ALCOHOL, MAGNESIUM ION, Spermidine N(1)-acetyltransferase | | 著者 | Osipiuk, J, VOLKART, L, MOY, S, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2015-07-17 | | 公開日 | 2015-07-29 | | 最終更新日 | 2019-12-11 | | 実験手法 | X-RAY DIFFRACTION (2.38 Å) | | 主引用文献 | Substrate-Induced Allosteric Change in the Quaternary Structure of the Spermidine N-Acetyltransferase SpeG.
J.Mol.Biol., 427, 2015
|
|
1YTL
 
 | | Crystal Structure of Acetyl-CoA decarboxylase/synthase complex epsilon subunit 2 | | 分子名称: | Acetyl-CoA decarboxylase/synthase complex epsilon subunit 2 | | 著者 | Chang, C, Evdokimova, E, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2005-02-10 | | 公開日 | 2005-03-22 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Crystal Structure of Acetyl-CoA decarboxylase/synthase complex epsilon subunit 2 from Archaeoglobus fulgidus
To be Published
|
|
