4R5Y
 
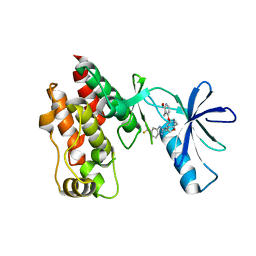 | | The complex structure of Braf V600E kinase domain with a novel Braf inhibitor | | 分子名称: | 5-({(1R,1aS,6bR)-1-[5-(trifluoromethyl)-1H-benzimidazol-2-yl]-1a,6b-dihydro-1H-cyclopropa[b][1]benzofuran-5-yl}oxy)-3,4-dihydro-1,8-naphthyridin-2(1H)-one, Serine/threonine-protein kinase B-raf | | 著者 | Feng, Y, Peng, H, Zhang, Y, Liu, Y, Wei, M. | | 登録日 | 2014-08-22 | | 公開日 | 2016-02-24 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (3.5 Å) | | 主引用文献 | BGB-283, a Novel RAF Kinase and EGFR Inhibitor, Displays Potent Antitumor Activity in BRAF-Mutated Colorectal Cancers.
Mol.Cancer Ther., 14, 2015
|
|
8G85
 
 | | vFP52.02 Fab in complex with BG505 DS-SOSIP Env trimer | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein gp120, ... | | 著者 | Gorman, J, Kwong, P.D. | | 登録日 | 2023-02-17 | | 公開日 | 2023-04-19 | | 最終更新日 | 2023-06-14 | | 実験手法 | ELECTRON MICROSCOPY (3.99 Å) | | 主引用文献 | Diverse Murine Vaccinations Reveal Distinct Antibody Classes to Target Fusion Peptide and Variation in Peptide Length to Improve HIV Neutralization.
J.Virol., 97, 2023
|
|
2VSL
 
 | |
3TL8
 
 | |
8G9W
 
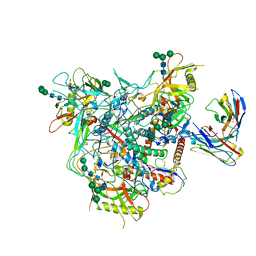 | | Cryo-EM structure of vFP49.02 Fab in complex with HIV-1 Env BG505 DS-SOSIP.664 (conformation 1) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein gp120, ... | | 著者 | Changela, A, Gorman, J, Kwong, P.D. | | 登録日 | 2023-02-22 | | 公開日 | 2023-04-19 | | 最終更新日 | 2023-06-14 | | 実験手法 | ELECTRON MICROSCOPY (4.66 Å) | | 主引用文献 | Diverse Murine Vaccinations Reveal Distinct Antibody Classes to Target Fusion Peptide and Variation in Peptide Length to Improve HIV Neutralization.
J.Virol., 97, 2023
|
|
8G9Y
 
 | | Cryo-EM structure of vFP49.02 Fab in complex with HIV-1 Env BG505 DS-SOSIP.664 (conformation 3) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein gp120, ... | | 著者 | Changela, A, Gorman, J, Kwong, P.D. | | 登録日 | 2023-02-22 | | 公開日 | 2023-04-19 | | 最終更新日 | 2023-06-14 | | 実験手法 | ELECTRON MICROSCOPY (4.28 Å) | | 主引用文献 | Diverse Murine Vaccinations Reveal Distinct Antibody Classes to Target Fusion Peptide and Variation in Peptide Length to Improve HIV Neutralization.
J.Virol., 97, 2023
|
|
8GAS
 
 | | vFP48.02 Fab in complex with BG505 DS-SOSIP Env trimer | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein gp120, ... | | 著者 | Gorman, J, Kwong, P.D. | | 登録日 | 2023-02-23 | | 公開日 | 2023-04-19 | | 最終更新日 | 2023-06-14 | | 実験手法 | ELECTRON MICROSCOPY (4.04 Å) | | 主引用文献 | Diverse Murine Vaccinations Reveal Distinct Antibody Classes to Target Fusion Peptide and Variation in Peptide Length to Improve HIV Neutralization.
J.Virol., 97, 2023
|
|
8G9X
 
 | | Cryo-EM structure of vFP49.02 Fab in complex with HIV-1 Env BG505 DS-SOSIP.664 (conformation 2) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein gp120, ... | | 著者 | Changela, A, Gorman, J, Kwong, P.D. | | 登録日 | 2023-02-22 | | 公開日 | 2023-04-19 | | 最終更新日 | 2023-06-14 | | 実験手法 | ELECTRON MICROSCOPY (4.46 Å) | | 主引用文献 | Diverse Murine Vaccinations Reveal Distinct Antibody Classes to Target Fusion Peptide and Variation in Peptide Length to Improve HIV Neutralization.
J.Virol., 97, 2023
|
|
6K2U
 
 | | Crystal structure of Thr66 ADP-ribosylated ubiquitin | | 分子名称: | ADENOSINE-5-DIPHOSPHORIBOSE, MAGNESIUM ION, Polyubiquitin-C, ... | | 著者 | Wang, X, Zhou, Y, Zhu, Y. | | 登録日 | 2019-05-15 | | 公開日 | 2020-03-18 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.554 Å) | | 主引用文献 | Threonine ADP-Ribosylation of Ubiquitin by a Bacterial Effector Family Blocks Host Ubiquitination.
Mol.Cell, 78, 2020
|
|
7V6A
 
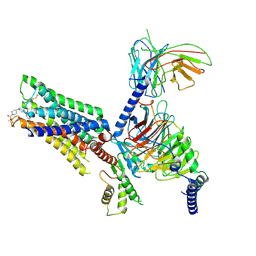 | | Cry-EM structure of M4-c110-G protein complex | | 分子名称: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | 著者 | Wang, J.J, Wu, M, Wu, L.J, Hua, T, Liu, Z.J, Wang, T. | | 登録日 | 2021-08-20 | | 公開日 | 2022-05-11 | | 最終更新日 | 2022-11-23 | | 実験手法 | ELECTRON MICROSCOPY (3.6 Å) | | 主引用文献 | The unconventional activation of the muscarinic acetylcholine receptor M4R by diverse ligands.
Nat Commun, 13, 2022
|
|
7V68
 
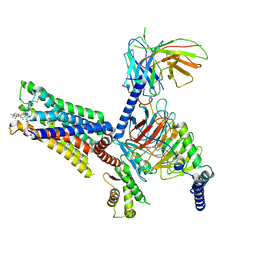 | | An Agonist and PAM-bound Class A GPCR with Gi protein complex structure | | 分子名称: | 3-amino-5-chloro-N-cyclopropyl-4-methyl-6-[2-(4-methylpiperazin-1-yl)-2-oxoethoxy]thieno[2,3-b]pyridine-2-carboxamide, 4-(4,5-dihydro-1,2-oxazol-3-yloxy)-N,N,N-trimethylbut-2-yn-1-aminium, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | 著者 | Wang, J.J, Wu, L.J, Wu, M, Hua, T, Liu, Z.J, Wang, T. | | 登録日 | 2021-08-20 | | 公開日 | 2022-05-11 | | 最終更新日 | 2022-11-23 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | The unconventional activation of the muscarinic acetylcholine receptor M4R by diverse ligands.
Nat Commun, 13, 2022
|
|
7V69
 
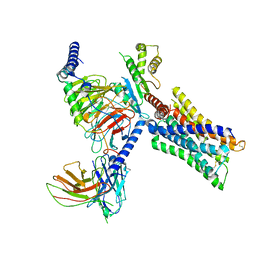 | | Cryo-EM structure of a class A GPCR-G protein complex | | 分子名称: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | 著者 | Wang, J.J, Wu, M, Wu, L.J, Hua, T, Liu, Z.J, Wang, T. | | 登録日 | 2021-08-20 | | 公開日 | 2022-05-11 | | 最終更新日 | 2022-11-23 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | The unconventional activation of the muscarinic acetylcholine receptor M4R by diverse ligands.
Nat Commun, 13, 2022
|
|
7LQH
 
 | | Cryo-EM of KFE8 thinner nanotube (class 2, 2-sub-1) | | 分子名称: | KFE8 peptide | | 著者 | Wang, F, Gnewou, O.M, Egelman, E.H, Conticello, V.P. | | 登録日 | 2021-02-13 | | 公開日 | 2021-06-30 | | 最終更新日 | 2023-11-15 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | Deterministic chaos in the self-assembly of beta sheet nanotubes from an amphipathic oligopeptide.
Matter, 4, 2021
|
|
7LQF
 
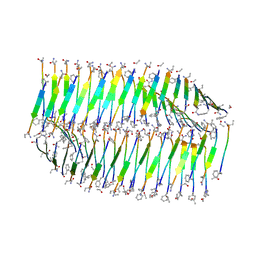 | |
7LQI
 
 | |
7LQE
 
 | |
7LQG
 
 | | Cryo-EM of the KFE8 thinner nanotube (class 1, C2) | | 分子名称: | KFE8 peptide | | 著者 | Wang, F, Gnewou, O.M, Egelman, E.H, Conticello, V.P. | | 登録日 | 2021-02-13 | | 公開日 | 2021-06-30 | | 最終更新日 | 2023-11-15 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | Deterministic chaos in the self-assembly of beta sheet nanotubes from an amphipathic oligopeptide.
Matter, 4, 2021
|
|
5SPY
 
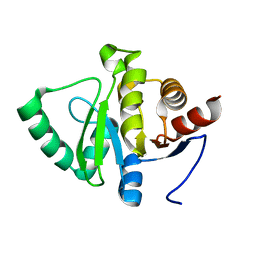 | |
5SQJ
 
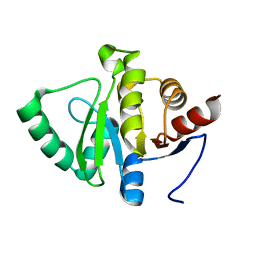 | |
5SQ8
 
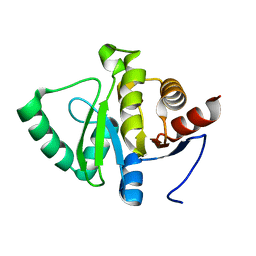 | |
5SQD
 
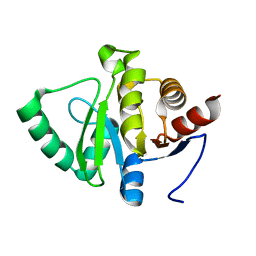 | | PanDDA analysis group deposition -- Crystal structure of SARS-CoV-2 NSP3 macrodomain in complex with Z5010894382 - (R,R) and (S,S) isomers | | 分子名称: | (1R,2R)-4-hydroxy-1-{[2-(hydroxymethyl)-1H-benzimidazole-5-carbonyl]amino}-2,3-dihydro-1H-indene-2-carboxylic acid, (1S,2S)-4-hydroxy-1-{[2-(hydroxymethyl)-1H-benzimidazole-5-carbonyl]amino}-2,3-dihydro-1H-indene-2-carboxylic acid, Non-structural protein 3 | | 著者 | Correy, G.J, Fraser, J.S. | | 登録日 | 2022-06-09 | | 公開日 | 2022-07-06 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.05 Å) | | 主引用文献 | Iterative computational design and crystallographic screening identifies potent inhibitors targeting the Nsp3 macrodomain of SARS-CoV-2.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
5SQP
 
 | |
5SQU
 
 | |
5SR7
 
 | | PanDDA analysis group deposition -- Crystal structure of SARS-CoV-2 NSP3 macrodomain in complex with Z4914649782 - (R,R,S) and (S,S,R) isomers | | 分子名称: | (1R,6S,7R)-3-(7-fluoro-9H-pyrimido[4,5-b]indol-4-yl)-3-azabicyclo[4.1.0]heptane-7-carboxylic acid, (1S,6R,7S)-3-(7-fluoro-9H-pyrimido[4,5-b]indol-4-yl)-3-azabicyclo[4.1.0]heptane-7-carboxylic acid, Non-structural protein 3 | | 著者 | Correy, G.J, Fraser, J.S. | | 登録日 | 2022-06-09 | | 公開日 | 2022-07-06 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.05 Å) | | 主引用文献 | Iterative computational design and crystallographic screening identifies potent inhibitors targeting the Nsp3 macrodomain of SARS-CoV-2.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
5SQ5
 
 | | PanDDA analysis group deposition -- Crystal structure of SARS-CoV-2 NSP3 macrodomain in complex with Z5010894407 - (R,S) and (S,S) isomers | | 分子名称: | (1R,2S)-1-[4-(cyclopropylcarbamamido)benzamido]-2,3-dihydro-1H-indene-2-carboxylic acid, (1S,2S)-1-[4-(cyclopropylcarbamamido)benzamido]-2,3-dihydro-1H-indene-2-carboxylic acid, Non-structural protein 3 | | 著者 | Correy, G.J, Fraser, J.S. | | 登録日 | 2022-06-09 | | 公開日 | 2022-07-06 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.05 Å) | | 主引用文献 | Iterative computational design and crystallographic screening identifies potent inhibitors targeting the Nsp3 macrodomain of SARS-CoV-2.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
