5JNH
 
 | |
2HOB
 
 | | Crystal structure of SARS-CoV main protease with authentic N and C-termini in complex with a Michael acceptor N3 | | 分子名称: | N-[(5-METHYLISOXAZOL-3-YL)CARBONYL]ALANYL-L-VALYL-N~1~-((1R,2Z)-4-(BENZYLOXY)-4-OXO-1-{[(3R)-2-OXOPYRROLIDIN-3-YL]METHYL}BUT-2-ENYL)-L-LEUCINAMIDE, Replicase polyprotein 1ab | | 著者 | Xue, X, Yang, H, Shen, W, Zhao, Q, Li, J, Rao, Z. | | 登録日 | 2006-07-14 | | 公開日 | 2007-04-03 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Production of authentic SARS-CoV M(pro) with enhanced activity: application as a novel tag-cleavage endopeptidase for protein overproduction
J.Mol.Biol., 366, 2007
|
|
6JIJ
 
 | |
6KW1
 
 | | The structure of the metallo-beta-lactamase VIM-2 in complex with a triazolylthioacetamide 1b | | 分子名称: | 2-[3-[2-(1H-benzimidazol-2-ylamino)-2-oxidanylidene-ethyl]sulfanyl-1H-1,2,4-triazol-5-yl]benzoic acid, ACETATE ION, Beta-lactamase class B VIM-2, ... | | 著者 | Yang, K.W, Xiang, Y. | | 登録日 | 2019-09-05 | | 公開日 | 2020-09-09 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.775212 Å) | | 主引用文献 | Kinetic, Thermodynamic, and Crystallographic Studies of 2-Triazolylthioacetamides as Verona Integron-Encoded Metallo-beta-Lactamase 2 (VIM-2) Inhibitor.
Biomolecules, 10, 2020
|
|
6MEV
 
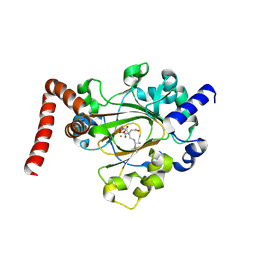 | | Structure of JMJD6 bound to Mono-Methyl Arginine. | | 分子名称: | (2S)-2-amino-5-[(N-methylcarbamimidoyl)amino]pentanoic acid, 2-OXOGLUTARIC ACID, Bifunctional arginine demethylase and lysyl-hydroxylase JMJD6, ... | | 著者 | Lee, S, Zhang, G. | | 登録日 | 2018-09-07 | | 公開日 | 2019-09-18 | | 最終更新日 | 2020-04-01 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | JMJD6 cleaves MePCE to release positive transcription elongation factor b (P-TEFb) in higher eukaryotes.
Elife, 9, 2020
|
|
8OTV
 
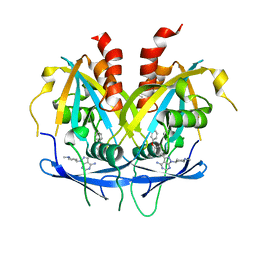 | | Crystal structure of NUDT14 complexed with novel compound | | 分子名称: | 1-(1-methylpiperidin-4-yl)-3-(4-phenoxyphenyl)pyrazolo[3,4-d]pyrimidin-4-amine, DIMETHYL SULFOXIDE, MAGNESIUM ION, ... | | 著者 | Balikci, E, Feyerherm, C, Bradshaw, W, Seupel, R, Brennan, P.E, Bountra, C, von Delft, F, Huber, K, Structural Genomics Consortium (SGC) | | 登録日 | 2023-04-21 | | 公開日 | 2024-05-01 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (1.82 Å) | | 主引用文献 | Unexpected Noncovalent Off-Target Activity of Clinical BTK Inhibitors Leads to Discovery of a Dual NUDT5/14 Antagonist.
J.Med.Chem., 67, 2024
|
|
7X4E
 
 | | Structure of 10635-DndE | | 分子名称: | DNA sulfur modification protein DndE, GLYCEROL | | 著者 | Haiyan, G, Wei, H, Chen, S, Wang, L, Wu, G. | | 登録日 | 2022-03-02 | | 公開日 | 2022-04-20 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (2.32 Å) | | 主引用文献 | Structural and Functional Analysis of DndE Involved in DNA Phosphorothioation in the Haloalkaliphilic Archaea Natronorubrum bangense JCM10635.
Mbio, 13, 2022
|
|
3OMG
 
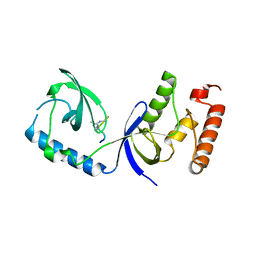 | | Structure of human SND1 extended tudor domain in complex with the symmetrically dimethylated arginine PIWIL1 peptide R14me2s | | 分子名称: | Staphylococcal nuclease domain-containing protein 1, dimethylated arginine peptide R14me2s | | 著者 | Lam, R, Liu, K, Guo, Y.H, Bian, C.B, Xu, C, MacKenzie, F, Bountra, C, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Min, J, Structural Genomics Consortium (SGC) | | 登録日 | 2010-08-26 | | 公開日 | 2010-09-08 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Structural basis for recognition of arginine methylated Piwi proteins by the extended Tudor domain.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
8OGX
 
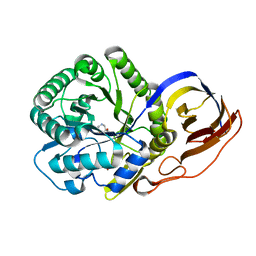 | | Beta-glucuronidase from Acidobacterium capsulatum in complex with inhibitor R3794 | | 分子名称: | (3~{S},4~{S})-4,5,5-tris(oxidanyl)piperidine-3-carboxylic acid, PHOSPHATE ION, beta-glucuronidase from Acidobacterium capsulatum | | 著者 | Moran, E.M, Davies, G.J, Chen, C, Nieuwendijk, E.V, Wu, L, Skoulikopoulou, F, Riet, V.V, Overkleeft, H.S, Armstrong, Z. | | 登録日 | 2023-03-20 | | 公開日 | 2024-01-10 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Molecular Basis for Inhibition of Heparanases and beta-Glucuronidases by Siastatin B.
J.Am.Chem.Soc., 146, 2024
|
|
3OMC
 
 | | Structure of human SND1 extended tudor domain in complex with the symmetrically dimethylated arginine PIWIL1 peptide R4me2s | | 分子名称: | CHLORIDE ION, SYNTHETIC PEPTIDE, Staphylococcal nuclease domain-containing protein 1 | | 著者 | Lam, R, Liu, K, Guo, Y.H, Bian, C.B, Xu, C, MacKenzie, F, Bountra, C, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Min, J, Structural Genomics Consortium (SGC) | | 登録日 | 2010-08-26 | | 公開日 | 2010-09-15 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.77 Å) | | 主引用文献 | Structural basis for recognition of arginine methylated Piwi proteins by the extended Tudor domain.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
7DD1
 
 | |
7DCD
 
 | |
8YZ7
 
 | |
7JT4
 
 | | Crystal Structure of BPTF bromodomain labelled with 5-fluoro-tryptophan | | 分子名称: | Nucleosome-remodeling factor subunit BPTF | | 著者 | Johnson, J.A, Shi, K, Aihara, H, Pomerantz, W.C.K. | | 登録日 | 2020-08-17 | | 公開日 | 2021-07-28 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.06 Å) | | 主引用文献 | Selectivity, ligand deconstruction, and cellular activity analysis of a BPTF bromodomain inhibitor
Org.Biomol.Chem., 17, 2019
|
|
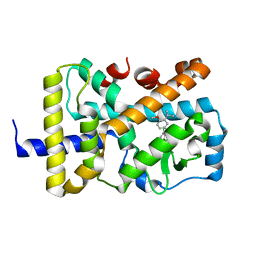
7KXD
 
 | |
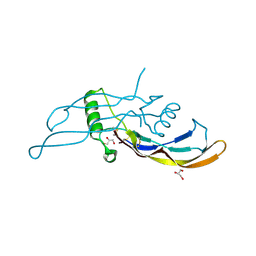
5I05
 
 | |
7W06
 
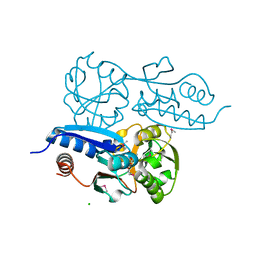 | | Itaconate inducible LysR-Type Transcriptional regulator (ITCR) in complex with itaconate (SeMet labeled), Space group C121. | | 分子名称: | 2-methylidenebutanedioic acid, CHLORIDE ION, SULFATE ION, ... | | 著者 | Sun, P.K, Wang, B, Wang, Z.X, Qi, S, Li, X.J. | | 登録日 | 2021-11-17 | | 公開日 | 2022-10-26 | | 最終更新日 | 2022-11-16 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | A genetically encoded fluorescent biosensor for detecting itaconate with subcellular resolution in living macrophages.
Nat Commun, 13, 2022
|
|
6NW9
 
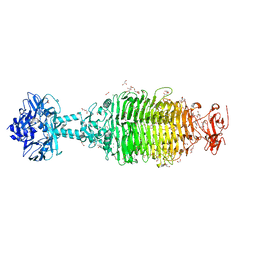 | | CRYSTAL STRUCTURE OF A TAILSPIKE PROTEIN 3 (TSP3, ORF212) FROM ESCHERICHIA COLI O157:H7 BACTERIOPHAGE CBA120 | | 分子名称: | 1,2-ETHANEDIOL, CARBONATE ION, CHLORIDE ION, ... | | 著者 | Greenfield, J.Y, Herzberg, O. | | 登録日 | 2019-02-06 | | 公開日 | 2019-06-05 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Structure and tailspike glycosidase machinery of ORF212 from E. coli O157:H7 phage CBA120 (TSP3).
Sci Rep, 9, 2019
|
|
7CMA
 
 | | Structure of A151R from African swine fever virus Georgia | | 分子名称: | A151R, ZINC ION | | 著者 | Niu, D, Liu, K, Huang, J, Chen, C, Liu, W, Guo, R. | | 登録日 | 2020-07-26 | | 公開日 | 2021-06-02 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (2.01 Å) | | 主引用文献 | Structure basis of non-structural protein pA151R from African Swine Fever Virus.
Biochem.Biophys.Res.Commun., 532, 2020
|
|
7X0L
 
 | |
7X0H
 
 | |
7X0P
 
 | |
7X0K
 
 | |
7X0G
 
 | |
7X0O
 
 | |
