6MEV
 
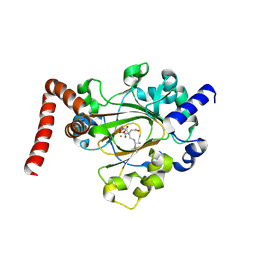 | | Structure of JMJD6 bound to Mono-Methyl Arginine. | | 分子名称: | (2S)-2-amino-5-[(N-methylcarbamimidoyl)amino]pentanoic acid, 2-OXOGLUTARIC ACID, Bifunctional arginine demethylase and lysyl-hydroxylase JMJD6, ... | | 著者 | Lee, S, Zhang, G. | | 登録日 | 2018-09-07 | | 公開日 | 2019-09-18 | | 最終更新日 | 2020-04-01 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | JMJD6 cleaves MePCE to release positive transcription elongation factor b (P-TEFb) in higher eukaryotes.
Elife, 9, 2020
|
|
8IYQ
 
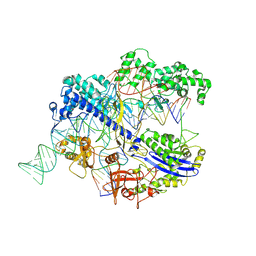 | | Structure of CbCas9 bound to 20-nucleotide complementary DNA substrate | | 分子名称: | NTS, TS, deadCbCas9, ... | | 著者 | Zhang, S, Lin, S, Liu, J.J.G. | | 登録日 | 2023-04-05 | | 公開日 | 2024-06-05 | | 最終更新日 | 2024-06-26 | | 実験手法 | ELECTRON MICROSCOPY (2.46 Å) | | 主引用文献 | Pro-CRISPR PcrIIC1-associated Cas9 system for enhanced bacterial immunity.
Nature, 630, 2024
|
|
2H2Z
 
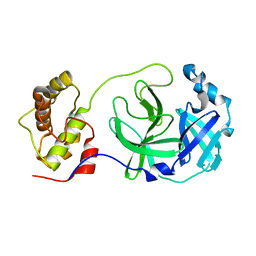 | | Crystal structure of SARS-CoV main protease with authentic N and C-termini | | 分子名称: | Replicase polyprotein 1ab | | 著者 | Yang, H, Xue, X, Shen, W, Zhao, Q, Rao, Z. | | 登録日 | 2006-05-20 | | 公開日 | 2007-04-03 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Production of authentic SARS-CoV M(pro) with enhanced activity: application as a novel tag-cleavage endopeptidase for protein overproduction
J.Mol.Biol., 366, 2007
|
|
2HOB
 
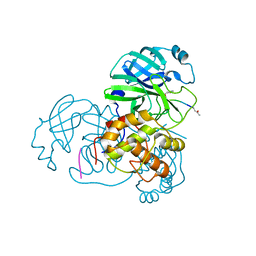 | | Crystal structure of SARS-CoV main protease with authentic N and C-termini in complex with a Michael acceptor N3 | | 分子名称: | N-[(5-METHYLISOXAZOL-3-YL)CARBONYL]ALANYL-L-VALYL-N~1~-((1R,2Z)-4-(BENZYLOXY)-4-OXO-1-{[(3R)-2-OXOPYRROLIDIN-3-YL]METHYL}BUT-2-ENYL)-L-LEUCINAMIDE, Replicase polyprotein 1ab | | 著者 | Xue, X, Yang, H, Shen, W, Zhao, Q, Li, J, Rao, Z. | | 登録日 | 2006-07-14 | | 公開日 | 2007-04-03 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Production of authentic SARS-CoV M(pro) with enhanced activity: application as a novel tag-cleavage endopeptidase for protein overproduction
J.Mol.Biol., 366, 2007
|
|
8GUG
 
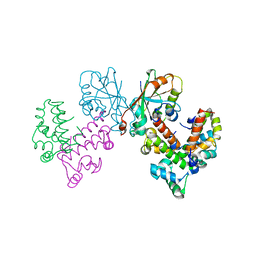 | | Structure of VPA0770 toxin bound to VPA0769 antitoxin in Vibrio parahaemolyticus | | 分子名称: | DUF2384 domain-containing protein, RES domain-containing protein | | 著者 | Song, X.J, Zhang, Y, Xu, Y.Y, Lin, Z. | | 登録日 | 2022-09-12 | | 公開日 | 2023-07-26 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (2.85 Å) | | 主引用文献 | Structural insights of the toxin-antitoxin system VPA0770-VPA0769 in Vibrio parahaemolyticus.
Int.J.Biol.Macromol., 242, 2023
|
|
7KXE
 
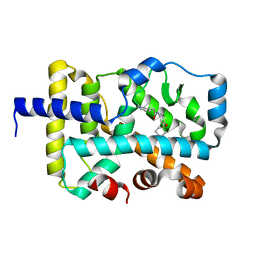 | |
7KXF
 
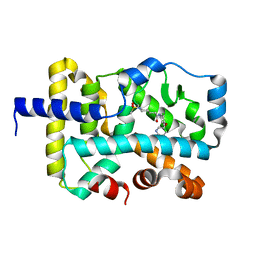 | |
7JT4
 
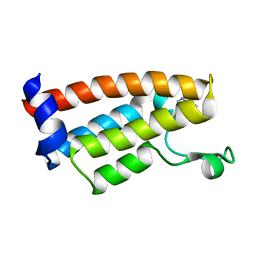 | | Crystal Structure of BPTF bromodomain labelled with 5-fluoro-tryptophan | | 分子名称: | Nucleosome-remodeling factor subunit BPTF | | 著者 | Johnson, J.A, Shi, K, Aihara, H, Pomerantz, W.C.K. | | 登録日 | 2020-08-17 | | 公開日 | 2021-07-28 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.06 Å) | | 主引用文献 | Selectivity, ligand deconstruction, and cellular activity analysis of a BPTF bromodomain inhibitor
Org.Biomol.Chem., 17, 2019
|
|
7KU4
 
 | |
7KWK
 
 | |
7KXD
 
 | |
6ISU
 
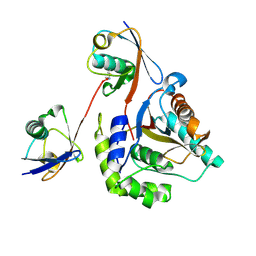 | | Crystal structure of Lys27-linked di-ubiquitin in complex with its selective interacting protein UCHL3 | | 分子名称: | Ubiquitin, Ubiquitin carboxyl-terminal hydrolase isozyme L3 | | 著者 | Ding, S, Pan, M, Zheng, Q, Ren, Y, Hong, D. | | 登録日 | 2018-11-19 | | 公開日 | 2019-02-06 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.866 Å) | | 主引用文献 | Chemical Protein Synthesis Enabled Mechanistic Studies on the Molecular Recognition of K27-linked Ubiquitin Chains.
Angew. Chem. Int. Ed. Engl., 58, 2019
|
|
6IVU
 
 | |
6IVS
 
 | |
2JWY
 
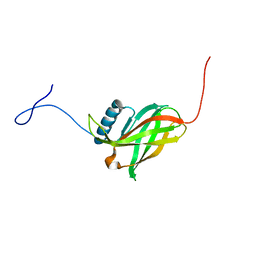 | | Solution NMR structure of uncharacterized lipoprotein yajI from Escherichia coli. Northeast Structural Genomics target ER540 | | 分子名称: | Uncharacterized lipoprotein yajI | | 著者 | Liu, G, Xiao, R, Chen, C, Liu, J, Baran, M.C, Swapna, G, Acton, T.B, Rost, B, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | 登録日 | 2007-10-31 | | 公開日 | 2007-11-20 | | 最終更新日 | 2024-05-08 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution NMR structure of uncharacterized lipoprotein yajI from Escherichia coli.
To be Published
|
|
5WJR
 
 | |
5X39
 
 | |
5X34
 
 | |
5XMK
 
 | | Cryo-EM structure of the ATP-bound Vps4 mutant-E233Q complex with Vta1 (masked) | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, Vacuolar protein sorting-associated protein 4, Vacuolar protein sorting-associated protein VTA1 | | 著者 | Sun, S, Li, L, Yang, F, Wang, X, Fan, F, Li, X, Wang, H, Sui, S. | | 登録日 | 2017-05-15 | | 公開日 | 2017-08-09 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (4.18 Å) | | 主引用文献 | Cryo-EM structures of the ATP-bound Vps4(E233Q) hexamer and its complex with Vta1 at near-atomic resolution
Nat Commun, 8, 2017
|
|
5XMI
 
 | | Cryo-EM Structure of the ATP-bound VPS4 mutant-E233Q hexamer (masked) | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, Vacuolar protein sorting-associated protein 4 | | 著者 | Sun, S, Li, L, Yang, F, Wang, X, Fan, F, Li, X, Wang, H, Sui, S. | | 登録日 | 2017-05-15 | | 公開日 | 2017-08-09 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (3.9 Å) | | 主引用文献 | Cryo-EM structures of the ATP-bound Vps4(E233Q) hexamer and its complex with Vta1 at near-atomic resolution
Nat Commun, 8, 2017
|
|
5X38
 
 | |
5X35
 
 | |
5X36
 
 | |
5X37
 
 | |
5X3C
 
 | |
