7ZDF
 
 | |
7ZDT
 
 | |
7XBK
 
 | | Structure and mechanism of a mitochondrial AAA+ disaggregase CLPB | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, Isoform 2 of Caseinolytic peptidase B protein homolog, MAGNESIUM ION, ... | | 著者 | Wu, D, Liu, Y, Dai, Y, Wang, G, Lu, G, Chen, Y, Li, N, Lin, J, Gao, N. | | 登録日 | 2022-03-21 | | 公開日 | 2023-01-25 | | 最終更新日 | 2024-06-26 | | 実験手法 | ELECTRON MICROSCOPY (3.7 Å) | | 主引用文献 | Comprehensive structural characterization of the human AAA+ disaggregase CLPB in the apo- and substrate-bound states reveals a unique mode of action driven by oligomerization.
Plos Biol., 21, 2023
|
|
7ZDE
 
 | |
7ZDL
 
 | |
7ZEC
 
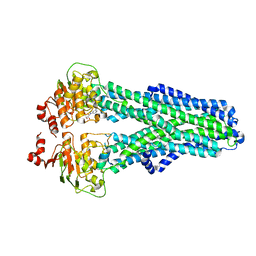 | |
2BKQ
 
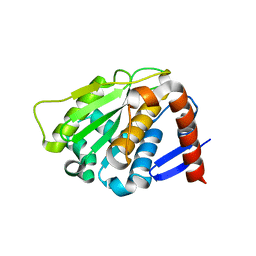 | | NEDD8 protease | | 分子名称: | SENTRIN-SPECIFIC PROTEASE 8 | | 著者 | Shen, L.N, Liu, H, Dong, C, Xirodimas, D, Naismith, J.H, Hay, R.T. | | 登録日 | 2005-02-18 | | 公開日 | 2005-02-21 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structural Basis of Nedd8 Ubiquitin Discrimination by the Deneddylating Enzyme Nedp1
Embo J., 24, 2005
|
|
8B30
 
 | |
2ZPS
 
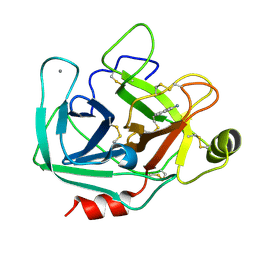 | |
8X2W
 
 | |
2C4N
 
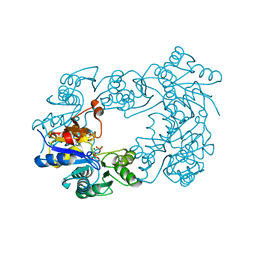 | | NagD from E.coli K-12 strain | | 分子名称: | MAGNESIUM ION, PHOSPHATE ION, PROTEIN NAGD | | 著者 | Tremblay, L.W, Dunaway-Mariano, D, Allen, K. | | 登録日 | 2005-10-20 | | 公開日 | 2006-01-26 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Structure and Activity Analyses of Escherichia Coli K-12 Nagd Provide Insight Into the Evolution of Biochemical Function in the Haloalkanoic Acid Dehalogenase Superfamily
Biochemistry, 45, 2006
|
|
2C4U
 
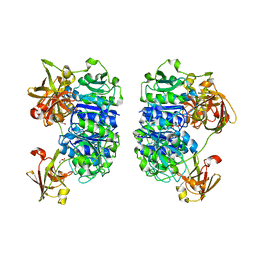 | | Crystal structure of the apo form of the 5'-Fluoro-5'-deoxyadenosine synthase enzyme from Streptomyces cattleya | | 分子名称: | 5'-FLUORO-5'-DEOXYADENOSINE SYNTHASE, GLYCEROL | | 著者 | McEwan, A.R, Deng, H, Robinson, D.A, DeLaurentis, W, McGlinchey, R.P, O'Hagan, D, Naismith, J.H. | | 登録日 | 2005-10-22 | | 公開日 | 2006-04-12 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Substrate specificity in enzymatic fluorination. The fluorinase from Streptomyces cattleya accepts 2'-deoxyadenosine substrates.
Org. Biomol. Chem., 4, 2006
|
|
2C5X
 
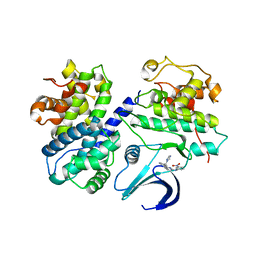 | | Differential Binding Of Inhibitors To Active And Inactive Cdk2 Provides Insights For Drug Design | | 分子名称: | CELL DIVISION PROTEIN KINASE 2, CYCLIN A2, HYDROXY(OXO)(3-{[(2Z)-4-[3-(1H-1,2,4-TRIAZOL-1-YLMETHYL)PHENYL]PYRIMIDIN-2(5H)-YLIDENE]AMINO}PHENYL)AMMONIUM | | 著者 | Kontopidis, G, Mcinnes, C, Pandalaneni, S.R, Mcnae, I, Gibson, D, Mezna, M, Thomas, M, Wood, G, Wang, S, Walkinshaw, M.D, Fischer, P.M. | | 登録日 | 2005-11-03 | | 公開日 | 2006-03-01 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Differential Binding of Inhibitors to Active and Inactive Cdk2 Provides Insights for Drug Design.
Chem.Biol., 13, 2006
|
|
3BGA
 
 | |
2BF0
 
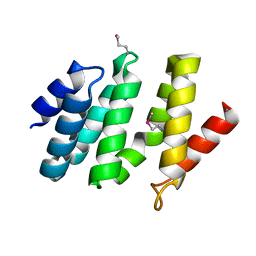 | | crystal structure of the rpr of pcf11 | | 分子名称: | CALCIUM ION, PCF11 | | 著者 | Noble, C.G, Hollingworth, D, Martin, S.R, Adeniran, V.E, Smerdon, S.J, Kelly, G, Taylor, I.A, Ramos, A. | | 登録日 | 2004-12-02 | | 公開日 | 2005-01-18 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Key Features of the Interaction between Pcf11 Cid and RNA Polymerase II Ctd.
Nat.Struct.Mol.Biol., 12, 2005
|
|
2BTQ
 
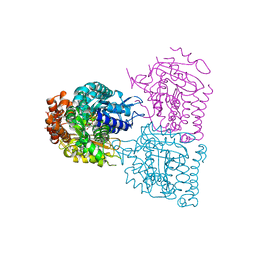 | |
2C1N
 
 | | Molecular basis for the recognition of phosphorylated and phosphoacetylated histone H3 by 14-3-3 | | 分子名称: | 14-3-3 PROTEIN ZETA/DELTA, HISTONE H3 ACETYLPHOSPHOPEPTIDE | | 著者 | Welburn, J.P.I, Macdonald, N, Noble, M.E.M, Nguyen, A, Yaffe, M.B, Clynes, D, Moggs, J.G, Orphanides, G, Thomson, S, Edmunds, J.W, Clayton, A.L, Endicott, J.A, Mahadevan, L.C. | | 登録日 | 2005-09-16 | | 公開日 | 2005-11-02 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Molecular Basis for the Recognition of Phosphorylated and Phosphoacetylated Histone H3 by 14-3-3.
Mol.Cell, 20, 2005
|
|
3BDU
 
 | | Crystal structure of protein Q6D8G1 at the resolution 1.9 A. Northeast Structural Genomics Consortium target EwR22A. | | 分子名称: | Putative lipoprotein | | 著者 | Kuzin, A.P, Su, M, Seetharaman, J, Wang, D, Fang, Y, Cunningham, K, Owens, L, Xiao, R, Liu, J, Baran, M.C, Acton, T.B, Rost, B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | 登録日 | 2007-11-15 | | 公開日 | 2007-11-27 | | 最終更新日 | 2019-12-25 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Crystal structure of protein Q6D8G1 at the resolution 1.9 A.
To be Published
|
|
2BN8
 
 | | Solution Structure and interactions of the E .coli Cell Division Activator Protein CedA | | 分子名称: | CELL DIVISION ACTIVATOR CEDA | | 著者 | Chen, H.A, Simpson, P, Huyton, T, Roper, D, Matthews, S. | | 登録日 | 2005-03-22 | | 公開日 | 2006-12-21 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution Structure and Interactions of the Escherichia Coli Cell Division Activator Protein Ceda.
Biochemistry, 44, 2005
|
|
8UXN
 
 | | Caulobacter crescentus FljM flagellar filament (symmetrized) | | 分子名称: | Flagellin FljM | | 著者 | Sanchez, J.C, Montemayor, E.J, Ploscariu, N.T, Parrell, D, Baumgardt, J.K, Yang, J.E, Sibert, B, Cai, K, Wright, E.R. | | 登録日 | 2023-11-09 | | 公開日 | 2024-06-26 | | 実験手法 | ELECTRON MICROSCOPY (2.11 Å) | | 主引用文献 | Direct evidence for multi-flagellin filament stabilization via atomic-level architecture of Caulobacter crescentus flagellar filaments
To Be Published
|
|
8UXJ
 
 | | Caulobacter crescentus FljK flagellar filament (asymmetrical) | | 分子名称: | Flagellin FljK | | 著者 | Sanchez, J.C, Montemayor, E.J, Ploscariu, N.T, Parrell, D, Baumgardt, J.K, Yang, J.E, Sibert, B, Cai, K, Wright, E.R. | | 登録日 | 2023-11-09 | | 公開日 | 2024-06-26 | | 実験手法 | ELECTRON MICROSCOPY (2.71 Å) | | 主引用文献 | Atomic-level architecture of Caulobacter crescentus flagellar filaments provide evidence for multi-flagellin filament stabilization
To Be Published
|
|
8VAC
 
 | | Cryogenic electron microscopy structure of human serum albumin in complex with teniposide | | 分子名称: | (5S,5aR,8aR,9R)-9-(4-hydroxy-3,5-dimethoxyphenyl)-8-oxo-5,5a,6,8,8a,9-hexahydrofuro[3',4':6,7]naphtho[2,3-d][1,3]dioxol -5-yl 4,6-O-(thiophen-2-ylmethylidene)-beta-D-glucopyranoside, Serum albumin | | 著者 | Catalano, C, Lucier, K.W, To, D, Senko, S, Tran, N.L, Farwell, A.C, Silva, S.M, Dip, P.V, Poweleit, N, Scapin, G. | | 登録日 | 2023-12-11 | | 公開日 | 2024-06-26 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | The CryoEM structure of human serum albumin in complex with ligands.
J.Struct.Biol., 216, 2024
|
|
3BHJ
 
 | | Crystal structure of human Carbonyl Reductase 1 in complex with glutathione | | 分子名称: | 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, 3-(4-AMINO-1-TERT-BUTYL-1H-PYRAZOLO[3,4-D]PYRIMIDIN-3-YL)PHENOL, Carbonyl reductase [NADPH] 1, ... | | 著者 | Rauh, D, Bateman, R.L, Shokat, K.M. | | 登録日 | 2007-11-28 | | 公開日 | 2008-10-21 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.77 Å) | | 主引用文献 | Human carbonyl reductase 1 is an s-nitrosoglutathione reductase
J.Biol.Chem., 283, 2008
|
|
2BKR
 
 | | NEDD8 NEDP1 complex | | 分子名称: | NEDDYLIN, SENTRIN-SPECIFIC PROTEASE 8 | | 著者 | Shen, L.N, Liu, H, Dong, C, Xirodimas, D, Naismith, J.H, Hay, R.T. | | 登録日 | 2005-02-18 | | 公開日 | 2005-09-15 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Structural Basis of Nedd8 Ubiquitin Discrimination by the Deneddylating Enzyme Nedp1
Embo J., 24, 2005
|
|
8VAE
 
 | | Cryogenic electron microscopy structure of human serum albumin in complex with salicylic acid | | 分子名称: | 2-HYDROXYBENZOIC ACID, Serum albumin | | 著者 | Catalano, C, Lucier, K.W, To, D, Senko, S, Tran, N.L, Farwell, A.C, Silva, S.M, Dip, P.V, Poweleit, N, Scapin, G. | | 登録日 | 2023-12-11 | | 公開日 | 2024-06-26 | | 実験手法 | ELECTRON MICROSCOPY (3.7 Å) | | 主引用文献 | The CryoEM structure of human serum albumin in complex with ligands.
J.Struct.Biol., 216, 2024
|
|
