5VD6
 
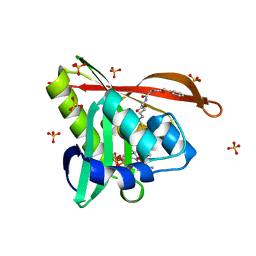 | | Crystal structure of a GNAT superfamily acetyltransferase PA4794 in complex with bisubstrate analog 6 | | 分子名称: | (3R,5S,9R,23S)-1-[(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-4-hydroxy-3-(phosphonooxy)tetrahydrofuran-2-yl]-3,5,9-trihydroxy-8,8-dimethyl-10,14-dioxo-23-({[(phenylacetyl)amino]acetyl}amino)-2,4,6-trioxa-18-thia-11,15-diaza-3,5-diphosphatetracosan-24-oic acid 3,5-dioxide (non-preferred name), SULFATE ION, acetyltransferase PA4794 | | 著者 | Majorek, K.A, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2017-04-01 | | 公開日 | 2017-07-26 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.2 Å) | | 主引用文献 | Generating enzyme and radical-mediated bisubstrates as tools for investigating Gcn5-related N-acetyltransferases.
FEBS Lett., 591, 2017
|
|
4N1X
 
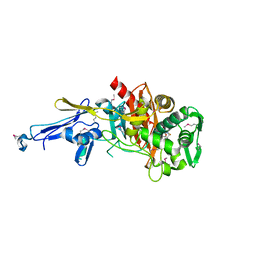 | | Structure of a putative peptidoglycan glycosyltransferase from Atopobium parvulum in complex with penicillin G | | 分子名称: | OPEN FORM - PENICILLIN G, Peptidoglycan glycosyltransferase | | 著者 | Filippova, E.V, Minasov, G, Shuvalova, L, Kiryukhina, O, Babnigg, G, Rubin, E, Sacchettini, J, Joachimiak, A, Anderson, W.F, Midwest Center for Structural Genomics (MCSG), Structures of Mtb Proteins Conferring Susceptibility to Known Mtb Inhibitors (MTBI) | | 登録日 | 2013-10-04 | | 公開日 | 2013-10-30 | | 最終更新日 | 2023-12-06 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structure of a putative peptidoglycan glycosyltransferase from Atopobium parvulum in complex with penicillin G
To be Published
|
|
3N3T
 
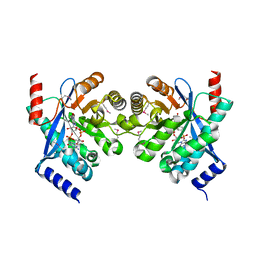 | | Crystal structure of putative diguanylate cyclase/phosphodiesterase complex with cyclic di-gmp | | 分子名称: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), CHLORIDE ION, MAGNESIUM ION, ... | | 著者 | Chang, C, Xu, X, Cui, H, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2010-05-20 | | 公開日 | 2010-06-16 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (2.35 Å) | | 主引用文献 | Structural insight into the mechanism of c-di-GMP hydrolysis by EAL domain phosphodiesterases.
J.Mol.Biol., 402, 2010
|
|
4R0J
 
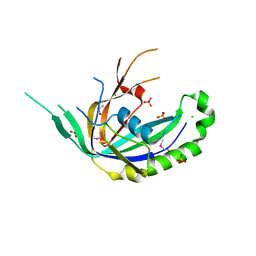 | | The crystal structure of a functionally uncharacterized protein SMU1763c from Streptococcus mutans | | 分子名称: | CHLORIDE ION, SULFATE ION, Uncharacterized protein | | 著者 | Tan, K, Xu, X, Cui, H, Liu, S, Savchenko, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2014-07-31 | | 公開日 | 2014-08-13 | | 実験手法 | X-RAY DIFFRACTION (1.715 Å) | | 主引用文献 | The crystal structure of a functionally uncharacterized protein SMU1763c from Streptococcus mutans
To be Published
|
|
4DQ0
 
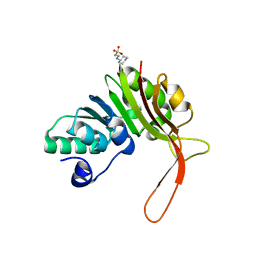 | |
4RBS
 
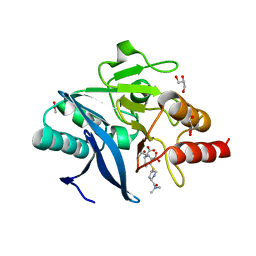 | | Crystal Structure of New Delhi Metallo-beta-Lactamase-1 in the Complex with Hydrolyzed Meropenem | | 分子名称: | (2S)-2-[(1S,2R)-1-carboxy-2-hydroxypropyl]-4-{[(3S,5S)-5-(dimethylcarbamoyl)pyrrolidin-3-yl]sulfanyl}-3-methyl-2H-pyrro le-5-carboxylic acid, ACETIC ACID, Beta-lactamase NDM-1, ... | | 著者 | Kim, Y, Tesar, C, Jedrzejczak, R, Babnigg, G, Sacchettini, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Structures of Mtb Proteins Conferring Susceptibility to Known Mtb Inhibitors (MTBI) | | 登録日 | 2014-09-12 | | 公開日 | 2014-11-26 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.405 Å) | | 主引用文献 | Crystal Structure of New Delhi Metallo-beta-Lactamase-1 in the Complex with Hydrolyzed Meropenem
To be Published
|
|
2GAU
 
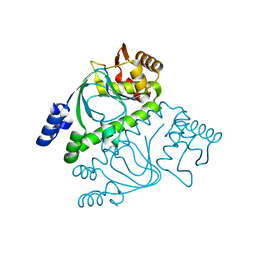 | | Crystal structure of transcriptional regulator, Crp/Fnr family from Porphyromonas gingivalis (APC80792), Structural genomics, MCSG | | 分子名称: | transcriptional regulator, Crp/Fnr family | | 著者 | Rotella, F.J, Zhang, R.G, Mulligan, R, Moy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2006-03-09 | | 公開日 | 2006-04-11 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | The 1.9-A crystal structure of transcriptional regulator, Crp/Fnr family from Porphyromonas gingivalis
To be Published
|
|
4RV5
 
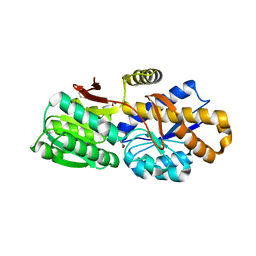 | | The crystal structure of a solute-binding protein from Anabaena variabilis ATCC 29413 in complex with pyruvic acid | | 分子名称: | Amino acid/amide ABC transporter substrate-binding protein, HAAT family, FORMIC ACID, ... | | 著者 | Tan, K, Li, H, Jedrzejczak, R, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2014-11-24 | | 公開日 | 2014-12-10 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.04 Å) | | 主引用文献 | The crystal structure of a solute-binding protein from Anabaena variabilis ATCC 29413 in complex with pyruvic acid
To be Published
|
|
3N2Q
 
 | |
4KUA
 
 | | Crystal structure of a GNAT superfamily acetyltransferase PA4794 | | 分子名称: | 1,2-ETHANEDIOL, GNAT superfamily acetyltransferase PA4794, SULFATE ION | | 著者 | Majorek, K.A, Chruszcz, M, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2013-05-21 | | 公開日 | 2013-06-05 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Structural, functional, and inhibition studies of a Gcn5-related N-acetyltransferase (GNAT) superfamily protein PA4794: a new C-terminal lysine protein acetyltransferase from pseudomonas aeruginosa.
J.Biol.Chem., 288, 2013
|
|
4PAG
 
 | | ABC transporter solute binding protein from Sulfurospirillum deleyianum DSM 6946 | | 分子名称: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, DI(HYDROXYETHYL)ETHER, HISTIDINE, ... | | 著者 | Chang, C, Endres, M, Li, H, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2014-04-08 | | 公開日 | 2014-04-30 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.901 Å) | | 主引用文献 | Crystal structure of ABC transporter solute binding protein from Sulfurospirillum deleyianum DSM 6946
To Be Published
|
|
4E5U
 
 | | The crystal structure of thymidylate kinase from Pseudomonas aeruginosa PAO1 in complex with thymidine monophosphate. | | 分子名称: | CALCIUM ION, THYMIDINE-5'-PHOSPHATE, Thymidylate kinase | | 著者 | Tan, K, Joachimiak, G, Jedrzejczak, R, Sacchettini, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Structures of Mtb Proteins Conferring Susceptibility to Known Mtb Inhibitors (MTBI) | | 登録日 | 2012-03-14 | | 公開日 | 2012-03-28 | | 実験手法 | X-RAY DIFFRACTION (1.812 Å) | | 主引用文献 | The crystal structure of thymidylate kinase from Pseudomonas aeruginosa PAO1 in complex with thymidine monophosphate.
To be Published
|
|
4KVF
 
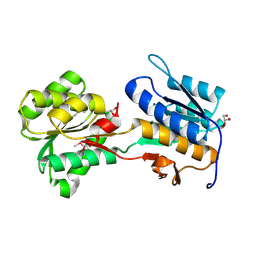 | | The crystal structure of a rhamnose ABC transporter, periplasmic rhamnose-binding protein from Kribbella flavida DSM 17836 | | 分子名称: | GLYCEROL, Rhamnose ABC transporter, periplasmic rhamnose-binding protein | | 著者 | Tan, K, Hatzos-Skintges, C, Endres, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2013-05-22 | | 公開日 | 2013-06-05 | | 実験手法 | X-RAY DIFFRACTION (1.722 Å) | | 主引用文献 | The crystal structure of a rhamnose ABC transporter, periplasmic rhamnose-binding protein from Kribbella flavida DSM 17836
To be Published
|
|
3OOU
 
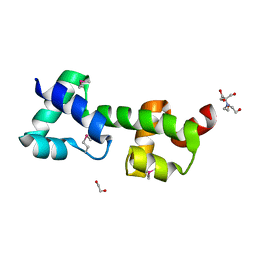 | | The structure of a protein with unkown function from Listeria innocua | | 分子名称: | 1,2-ETHANEDIOL, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Lin2118 protein | | 著者 | Fan, Y, Mack, J, Feldman, B, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2010-08-31 | | 公開日 | 2010-09-22 | | 最終更新日 | 2017-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.57 Å) | | 主引用文献 | The structure of a protein with unkown function from Listeria innocua
To be Published
|
|
4Q6T
 
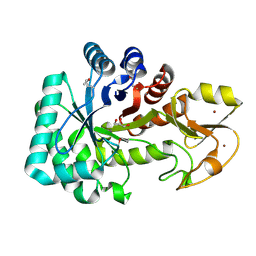 | | The crystal structure of a class V chitininase from Pseudomonas fluorescens Pf-5 | | 分子名称: | CADMIUM ION, CHLORIDE ION, GLYCEROL, ... | | 著者 | Tan, K, Mack, J.C, Endres, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2014-04-23 | | 公開日 | 2014-05-07 | | 最終更新日 | 2017-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | The crystal structure of a class V chitininase from Pseudomonas fluorescens Pf-5
To be Published
|
|
5VES
 
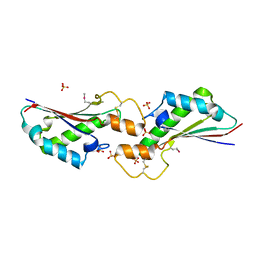 | | The 2.4A crystal structure of OmpA domain of OmpA from Salmonella enterica subsp. enterica serovar Typhimurium str. 14028S | | 分子名称: | Outer membrane protein A, SULFATE ION | | 著者 | Tan, K, Wu, R, Jedrzejczak, R, Adkins, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Program for the Characterization of Secreted Effector Proteins (PCSEP) | | 登録日 | 2017-04-05 | | 公開日 | 2017-04-19 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Insights into PG-binding, conformational change, and dimerization of the OmpA C-terminal domains from Salmonella enterica serovar Typhimurium and Borrelia burgdorferi.
Protein Sci., 26, 2017
|
|
4RW0
 
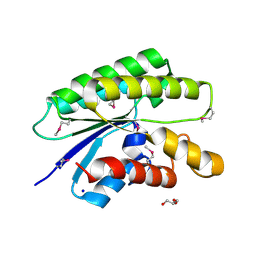 | | Crystal structure of a member of the lipolytic protein G-D-S-L family from Veillonella parvula DSM 2008 | | 分子名称: | GLYCEROL, Lipolytic protein G-D-S-L family, SODIUM ION | | 著者 | Nocek, B, Hatzos-Skintges, C, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2014-11-30 | | 公開日 | 2015-01-28 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Crystal structure of a member of the lipolytic protein G-D-S-L family from Veillonella parvula DSM 2008
To be Published
|
|
4MV2
 
 | | Crystal structure of plu4264 protein from Photorhabdus luminescens | | 分子名称: | NICKEL (II) ION, SODIUM ION, plu4264 | | 著者 | Michalska, K, Li, H, Jedrzejczak, R, Babnigg, G, Bingman, C.A, Yennamalli, R, Weerth, S, Thomas, M.G, Phillips Jr, G.N, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | 登録日 | 2013-09-23 | | 公開日 | 2013-10-02 | | 最終更新日 | 2015-02-04 | | 実験手法 | X-RAY DIFFRACTION (1.349 Å) | | 主引用文献 | Structure of a cupin protein Plu4264 from Photorhabdus luminescens subsp. laumondii TTO1 at 1.35 angstrom resolution.
Proteins, 83, 2015
|
|
4R23
 
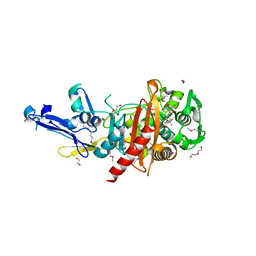 | | Structure of a putative peptidoglycan glycosyltransferase from Atopobium parvulum in complex with dicloxacillin | | 分子名称: | (3R,4R,5R)-3-(2,6-dichlorophenyl)-N-{(1R)-1-[(2R,4S)-4-(dihydroxymethyl)-5,5-dimethyl-1,3-thiazolidin-2-yl]-2-oxoethyl} -5-methyl-1,2-oxazolidine-4-carboxamide, 1,2-ETHANEDIOL, 1-ETHOXY-2-(2-ETHOXYETHOXY)ETHANE, ... | | 著者 | Filippova, E.V, Minasov, G, Kiryukhina, O, Clancy, S, Joachimiak, A, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2014-08-08 | | 公開日 | 2014-09-17 | | 最終更新日 | 2023-12-06 | | 実験手法 | X-RAY DIFFRACTION (1.84 Å) | | 主引用文献 | Structure of a putative peptidoglycan glycosyltransferase from Atopobium parvulum in complex with dicloxacillin
To be Published
|
|
4RDC
 
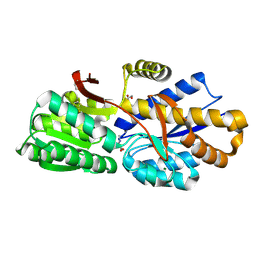 | | The crystal structure of a solute-binding protein (N280D mutant) from Anabaena variabilis ATCC 29413 in complex with proline | | 分子名称: | Amino acid/amide ABC transporter substrate-binding protein, HAAT family, FORMIC ACID, ... | | 著者 | Tan, K, Li, H, Jedrzejczak, R, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2014-09-18 | | 公開日 | 2014-10-01 | | 最終更新日 | 2024-10-09 | | 実験手法 | X-RAY DIFFRACTION (1.198 Å) | | 主引用文献 | The crystal structure of a solute-binding protein (N280D mutant) from Anabaena variabilis ATCC 29413 in complex with proline.
To be Published
|
|
4RWU
 
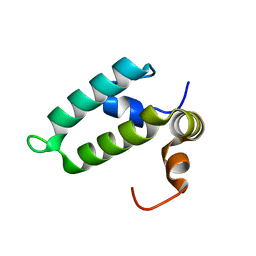 | | J-domain of Sis1 protein, Hsp40 co-chaperone from Saccharomyces cerevisiae | | 分子名称: | Protein SIS1 | | 著者 | Osipiuk, J, Zhou, M, Gu, M, Sahi, C, Craig, E.A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2014-12-05 | | 公開日 | 2014-12-17 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.25 Å) | | 主引用文献 | Roles of intramolecular and intermolecular interactions in functional regulation of the Hsp70 J-protein co-chaperone Sis1.
J.Mol.Biol., 427, 2015
|
|
4U28
 
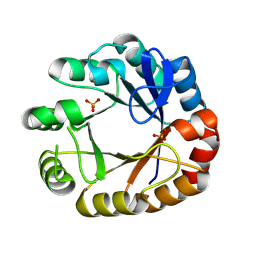 | | Crystal structure of apo Phosphoribosyl isomerase A from Streptomyces sviceus ATCC 29083 | | 分子名称: | PHOSPHATE ION, Phosphoribosyl isomerase A | | 著者 | Chang, C, Verduzco-Castro, E.A, Endres, M, Barona-Gomez, F, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2014-07-16 | | 公開日 | 2014-07-30 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.33 Å) | | 主引用文献 | Co-occurrence of analogous enzymes determines evolution of a novel ( beta alpha )8-isomerase sub-family after non-conserved mutations in flexible loop.
Biochem. J., 473, 2016
|
|
1ILV
 
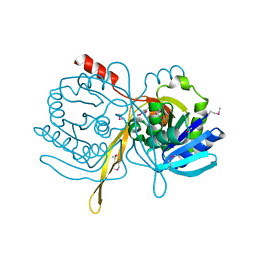 | | Crystal Structure Analysis of the TM107 | | 分子名称: | STATIONARY-PHASE SURVIVAL PROTEIN SURE HOMOLOG | | 著者 | Zhang, R, Joachimiak, A, Edwards, A, Savchenko, A, Beasley, S, Evdokimova, E, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2001-05-08 | | 公開日 | 2001-10-10 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structure of Thermotoga maritima stationary phase survival protein SurE: a novel acid phosphatase.
Structure, 9, 2001
|
|
3MOI
 
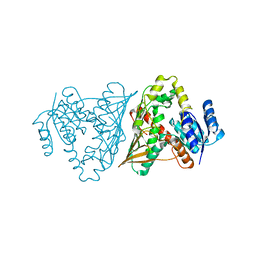 | | The crystal structure of the putative dehydrogenase from Bordetella bronchiseptica RB50 | | 分子名称: | Probable dehydrogenase | | 著者 | Zhang, R, Evdokimova, E, Egorova, O, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2010-04-22 | | 公開日 | 2010-05-26 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | The crystal structure of the putative dehydrogenase from Bordetella bronchiseptica RB50
To be Published
|
|
3M33
 
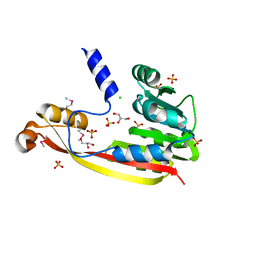 | | The crystal structure of a functionally unknown protein from Deinococcus radiodurans R1 | | 分子名称: | CHLORIDE ION, GLYCEROL, SULFATE ION, ... | | 著者 | Tan, K, Mack, J, Feldmann, B, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2010-03-08 | | 公開日 | 2010-03-31 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (2.195 Å) | | 主引用文献 | The crystal structure of a functionally unknown protein from Deinococcus radiodurans R1
To be Published
|
|
