8BIC
 
 | |
8BIR
 
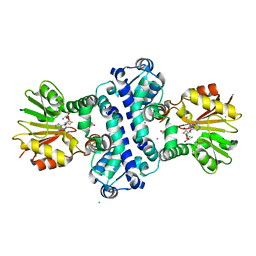 | | O-Methyltransferase Plu4895 in complex with SAH and AQ-256 | | 分子名称: | 1,3,8-tris(oxidanyl)anthracene-9,10-dione, CHLORIDE ION, IODIDE ION, ... | | 著者 | Huber, E.M, Groll, M. | | 登録日 | 2022-11-02 | | 公開日 | 2023-03-08 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | A set of closely related methyltransferases for site-specific tailoring of anthraquinone pigments.
Structure, 31, 2023
|
|
8BGZ
 
 | | O-Methyltransferase Plu4890 (mutant H229N) in complex with SAH and AQ-256 | | 分子名称: | 1,3,8-tris(oxidanyl)anthracene-9,10-dione, GLYCEROL, S-ADENOSYL-L-HOMOCYSTEINE, ... | | 著者 | Huber, E.M, Groll, M. | | 登録日 | 2022-10-28 | | 公開日 | 2023-03-08 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | A set of closely related methyltransferases for site-specific tailoring of anthraquinone pigments.
Structure, 31, 2023
|
|
8BGX
 
 | | O-Methyltransferase Plu4890 in complex with SAH and AQ-270a | | 分子名称: | 1-methoxy-3,8-bis(oxidanyl)anthracene-9,10-dione, CHLORIDE ION, GLYCEROL, ... | | 著者 | Huber, E.M, Groll, M. | | 登録日 | 2022-10-28 | | 公開日 | 2023-03-08 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | A set of closely related methyltransferases for site-specific tailoring of anthraquinone pigments.
Structure, 31, 2023
|
|
8BIH
 
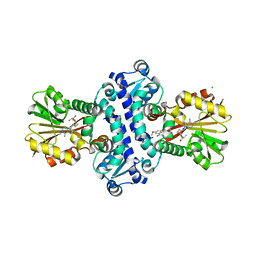 | | O-Methyltransferase Plu4890 in complex with SAH and AQ-284b | | 分子名称: | 3,8-dimethoxy-1-oxidanyl-anthracene-9,10-dione, CHLORIDE ION, GLYCEROL, ... | | 著者 | Huber, E.M, Groll, M. | | 登録日 | 2022-11-02 | | 公開日 | 2023-03-08 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | A set of closely related methyltransferases for site-specific tailoring of anthraquinone pigments.
Structure, 31, 2023
|
|
8BIF
 
 | |
8BID
 
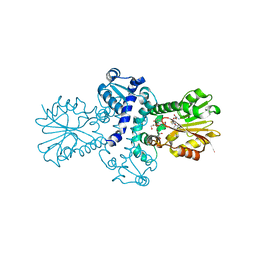 | | O-Methyltransferase Plu4890 (mutant H229N) in complex with SAH and AQ-270a | | 分子名称: | 1-methoxy-3,8-bis(oxidanyl)anthracene-9,10-dione, CHLORIDE ION, GLYCEROL, ... | | 著者 | Huber, E.M, Groll, M. | | 登録日 | 2022-11-02 | | 公開日 | 2023-03-08 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | A set of closely related methyltransferases for site-specific tailoring of anthraquinone pigments.
Structure, 31, 2023
|
|
4FEH
 
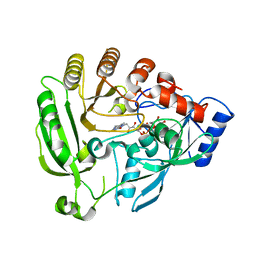 | |
4FF6
 
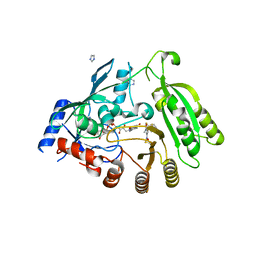 | | Mycobacterium tuberculosis DprE1 in complex with CT325 - monoclinic crystal form | | 分子名称: | 3-(hydroxyamino)-N-[(1R)-1-phenylethyl]-5-(trifluoromethyl)benzamide, FLAVIN-ADENINE DINUCLEOTIDE, IMIDAZOLE, ... | | 著者 | Batt, S.M, Besra, G.S, Futterer, K. | | 登録日 | 2012-05-31 | | 公開日 | 2012-07-04 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Structural basis of inhibition of Mycobacterium tuberculosis DprE1 by benzothiazinone inhibitors.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4CD0
 
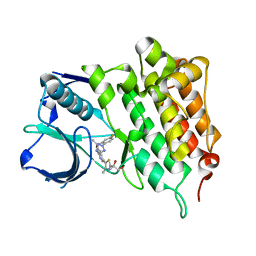 | | Structure of L1196M Mutant Human Anaplastic Lymphoma Kinase in Complex with 2-(5-(6-amino-5-((R)-1-(5-fluoro-2-(2H-1,2,3-triazol-2- yl)phenyl)ethoxy)pyridin-3-yl)-4-methylthiazol-2-yl)propane-1,2-diol | | 分子名称: | (2R)-2-[5-(6-amino-5-{(1R)-1-[2-(1,3-dihydro-2H-1,2,3-triazol-2-yl)-5-fluorophenyl]ethoxy}pyridin-3-yl)-4-methyl-1,3-thiazol-2-yl]propane-1,2-diol, ALK TYROSINE KINASE RECEPTOR | | 著者 | McTigue, M, Deng, Y, Liu, W, Brooun, A, Stewart, A. | | 登録日 | 2013-10-29 | | 公開日 | 2014-01-29 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.23 Å) | | 主引用文献 | The Design of Potent and Selective Inhibitors to Overcome Clinical Alk Mutations Resistant to Crizotinib.
J.Med.Chem., 57, 2014
|
|
4CNH
 
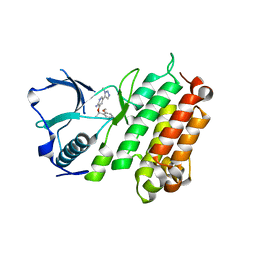 | | Structure of the Human Anaplastic Lymphoma Kinase in Complex with the inhibitor 3-((1R)-1-(5-fluoro-2-methoxyphenyl)ethoxy)-5-(1-methyl-1H- 1,2,3-triazol-5-yl)pyridin-2-amine | | 分子名称: | 3-[(1R)-1-(5-fluoro-2-methoxyphenyl)ethoxy]-5-(1-methyl-1H-1,2,3-triazol-5-yl)pyridin-2-amine, ALK TYROSINE KINASE RECEPTOR | | 著者 | McTigue, M.A, Deng, Y.L, Liu, W, Brooun, A, Stewart, A.E. | | 登録日 | 2014-01-22 | | 公開日 | 2014-05-28 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Discovery of (10R)-7-Amino-12-Fluoro-2,10,16-Trimethyl-15-Oxo-10,15,16,17-Tetrahydro-2H-8,4-(Metheno)Pyrazolo[4,3-H][2,5,11]Benzoxadiazacyclotetradecine-3-Carbonitrile (Pf-06463922), a Macrocyclic Inhibitor of Alk/Ros1 with Pre-Clinical Brain Exposure and Broad Spectrum Potency Against Alk-Resistant Mutations.
J.Med.Chem., 57, 2014
|
|
4CLI
 
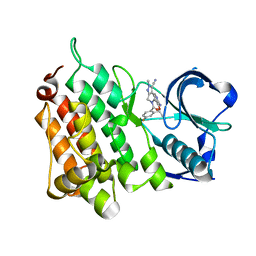 | | Structure of the Human Anaplastic Lymphoma Kinase in Complex with PF- 06463922 ((10R)-7-amino-12-fluoro-2,10,16-trimethyl-15-oxo-10,15,16, 17-tetrahydro-2H-8,4-(metheno)pyrazolo(4,3-h)(2,5,11) benzoxadiazacyclotetradecine-3-carbonitrile). | | 分子名称: | (10R)-7-amino-12-fluoro-2,10,16-trimethyl-15-oxo-10,15,16,17-tetrahydro-2H-8,4-(metheno)pyrazolo[4,3-h][2,5,11]benzoxadiazacyclotetradecine-3-carbonitrile, ALK TYROSINE KINASE RECEPTOR | | 著者 | McTigue, M.A, Deng, Y.L, Liu, W, Brooun, A, Stewart, A.E. | | 登録日 | 2014-01-14 | | 公開日 | 2014-05-28 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.05 Å) | | 主引用文献 | Discovery of (10R)-7-Amino-12-Fluoro-2,10,16-Trimethyl-15-Oxo-10,15,16,17-Tetrahydro-2H-8,4-(Metheno)Pyrazolo[4,3-H][2,5,11]Benzoxadiazacyclotetradecine-3-Carbonitrile (Pf-06463922), a Macrocyclic Inhibitor of Alk/Ros1 with Pre-Clinical Brain Exposure and Broad Spectrum Potency Against Alk-Resistant Mutations.
J.Med.Chem., 57, 2014
|
|
4CMT
 
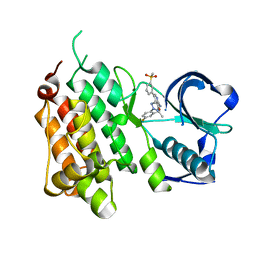 | | Structure of the Human Anaplastic Lymphoma Kinase in Complex with the inhibitor 3-((1R)-1-(5-fluoro-2-(2H-1,2,3-triazol-2-yl)phenyl)ethoxy)- 5-(3-(methylsulfonyl)phenyl)pyridin-2-amine | | 分子名称: | 3-{(1R)-1-[2-(1,3-dihydro-2H-1,2,3-triazol-2-yl)-5-fluorophenyl]ethoxy}-5-[3-(methylsulfonyl)phenyl]pyridin-2-amine, ALK TYROSINE KINASE RECEPTOR | | 著者 | McTigue, M.A, Deng, Y.L, Liu, W, Brooun, A, Stewart, A.E. | | 登録日 | 2014-01-17 | | 公開日 | 2014-05-28 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.73 Å) | | 主引用文献 | Discovery of (10R)-7-Amino-12-Fluoro-2,10,16-Trimethyl-15-Oxo-10,15,16,17-Tetrahydro-2H-8,4-(Metheno)Pyrazolo[4,3-H][2,5,11]Benzoxadiazacyclotetradecine-3-Carbonitrile (Pf-06463922), a Macrocyclic Inhibitor of Alk/Ros1 with Pre-Clinical Brain Exposure and Broad Spectrum Potency Against Alk-Resistant Mutations.
J.Med.Chem., 57, 2014
|
|
4CMO
 
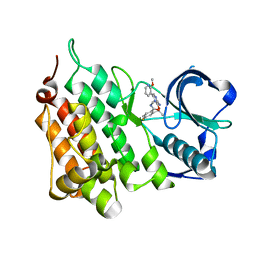 | | Structure of the Human Anaplastic Lymphoma Kinase in Complex with the inhibitor 2-((1R)-1-((3-amino-6-(2-methoxypyridin-3-yl)pyrazin-2-yl) oxy)ethyl)-4-fluoro-N-methylbenzamide | | 分子名称: | 2-[(1R)-1-{[3-amino-6-(2-methoxypyridin-3-yl)pyrazin-2-yl]oxy}ethyl]-4-fluoro-N-methylbenzamide, ALK TYROSINE KINASE RECEPTOR | | 著者 | McTigue, M.A, Deng, Y.L, Liu, W, Brooun, A, Stewart, A.E. | | 登録日 | 2014-01-16 | | 公開日 | 2014-05-28 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.05 Å) | | 主引用文献 | Discovery of (10R)-7-Amino-12-Fluoro-2,10,16-Trimethyl-15-Oxo-10,15,16,17-Tetrahydro-2H-8,4-(Metheno)Pyrazolo[4,3-H][2,5,11]Benzoxadiazacyclotetradecine-3-Carbonitrile (Pf-06463922), a Macrocyclic Inhibitor of Alk/Ros1 with Pre-Clinical Brain Exposure and Broad Spectrum Potency Against Alk-Resistant Mutations.
J.Med.Chem., 57, 2014
|
|
4CMU
 
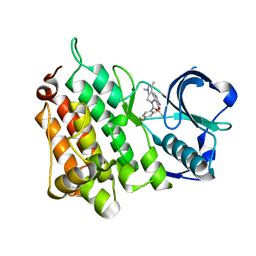 | | Structure of the Human Anaplastic Lymphoma Kinase in Complex with the inhibitor (10R)-7-amino-12-fluoro-1,3,10,16-tetramethyl-16,17-dihydro- 1H-8,4-(metheno)pyrazolo(4,3-h)(2,5,11)benzoxadiazacyclotetradecin-15(10H)-one | | 分子名称: | (10R)-7-amino-12-fluoro-1,3,10,16-tetramethyl-16,17-dihydro-1H-8,4-(metheno)pyrazolo[4,3-h][2,5,11]benzoxadiazacyclotetradecin-15(10H)-one, ALK TYROSINE KINASE RECEPTOR | | 著者 | McTigue, M.A, Deng, Y.L, Liu, W, Brooun, A, Stewart, A.E. | | 登録日 | 2014-01-17 | | 公開日 | 2014-05-28 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Discovery of (10R)-7-Amino-12-Fluoro-2,10,16-Trimethyl-15-Oxo-10,15,16,17-Tetrahydro-2H-8,4-(Metheno)Pyrazolo[4,3-H][2,5,11]Benzoxadiazacyclotetradecine-3-Carbonitrile (Pf-06463922), a Macrocyclic Inhibitor of Alk/Ros1 with Pre-Clinical Brain Exposure and Broad Spectrum Potency Against Alk-Resistant Mutations.
J.Med.Chem., 57, 2014
|
|
4CTC
 
 | | Structure of the Human Anaplastic Lymphoma Kinase in Complex with the inhibitor 7-amino-3-cyclopropyl-12-fluoro-1,10,16-trimethyl-16,17- dihydro-1H-8,4-(metheno)pyrazolo(4,3-h)(2,5,11) benzoxadiazacyclotetradecin-15(10H)-one | | 分子名称: | (10R)-7-amino-3-cyclopropyl-12-fluoro-1,10,16-trimethyl-16,17-dihydro-1H-8,4-(metheno)pyrazolo[4,3-h][2,5,11]benzoxadiazacyclotetradecin-15(10H)-one, ALK TYROSINE KINASE RECEPTOR | | 著者 | McTigue, M.A, Deng, Y.L, Liu, W, Brooun, A, Stewart, A.E. | | 登録日 | 2014-03-12 | | 公開日 | 2014-05-28 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.03 Å) | | 主引用文献 | Discovery of (10R)-7-Amino-12-Fluoro-2,10,16-Trimethyl-15-Oxo-10,15,16,17-Tetrahydro-2H-8,4-(Metheno)Pyrazolo[4,3-H][2,5,11]Benzoxadiazacyclotetradecine-3-Carbonitrile (Pf-06463922), a Macrocyclic Inhibitor of Alk/Ros1 with Pre-Clinical Brain Exposure and Broad Spectrum Potency Against Alk-Resistant Mutations.
J.Med.Chem., 57, 2014
|
|
2DTJ
 
 | | Crystal structure of regulatory subunit of aspartate kinase from Corynebacterium glutamicum | | 分子名称: | Aspartokinase, CITRIC ACID, THREONINE | | 著者 | Yoshida, A, Tomita, T, Fushinobu, S, Kuzuyama, T, Nishiyama, M. | | 登録日 | 2006-07-12 | | 公開日 | 2007-05-01 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.58 Å) | | 主引用文献 | Structural Insight into Concerted Inhibition of alpha(2)beta(2)-Type Aspartate Kinase from Corynebacterium glutamicum
J.Mol.Biol., 368, 2007
|
|
4PXH
 
 | |
4PWV
 
 | |
8UAJ
 
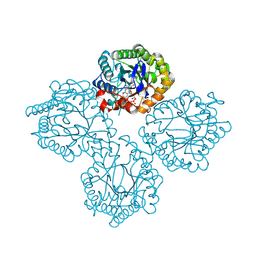 | | Succinate Bound Crystal Structure of Thermus scotoductus SA-01 Ene-reductase | | 分子名称: | FLAVIN MONONUCLEOTIDE, NADPH dehydrogenase, SUCCINIC ACID | | 著者 | Wilson, L.A, Guddat, L, Schenk, G, Scott, C. | | 登録日 | 2023-09-21 | | 公開日 | 2024-07-03 | | 実験手法 | X-RAY DIFFRACTION (2.14 Å) | | 主引用文献 | Structural Characterization of Enzymatic Interactions with Functional Nicotinamide Cofactor Biomimetics
Catalysts, 14, 2024
|
|
8VCC
 
 | |
4CTB
 
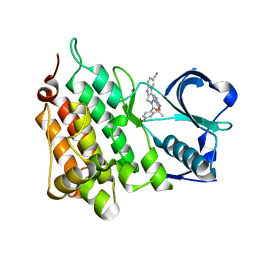 | | Structure of the Human Anaplastic Lymphoma Kinase in Complex with the inhibitor (5R)-8-amino-3-fluoro-5,19-dimethyl-20-oxo-5,18,19,20- tetrahydro-7,11-(azeno)pyrido(2',1':2,3)imidazo(4,5-h)(2,5,11) benzoxadiazacyclotetradecine-14-carbonitrile | | 分子名称: | (5R)-8-amino-3-fluoro-5,19-dimethyl-20-oxo-5,18,19,20-tetrahydro-11,7-(azeno)pyrido[2',1':2,3]imidazo[4,5-h][2,5,11]benzoxadiazacyclotetradecine-14-carbonitrile, ALK TYROSINE KINASE RECEPTOR | | 著者 | McTigue, M.A, Deng, Y.L, Liu, W, Brooun, A, Stewart, A.E. | | 登録日 | 2014-03-12 | | 公開日 | 2014-05-28 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.79 Å) | | 主引用文献 | Discovery of (10R)-7-Amino-12-Fluoro-2,10,16-Trimethyl-15-Oxo-10,15,16,17-Tetrahydro-2H-8,4-(Metheno)Pyrazolo[4,3-H][2,5,11]Benzoxadiazacyclotetradecine-3-Carbonitrile (Pf-06463922), a Macrocyclic Inhibitor of Alk/Ros1 with Pre-Clinical Brain Exposure and Broad Spectrum Potency Against Alk-Resistant Mutations.
J.Med.Chem., 57, 2014
|
|
4CLJ
 
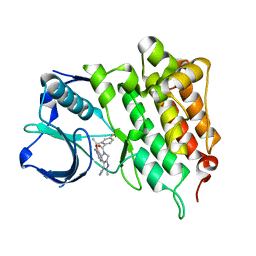 | | Structure of L1196M Mutant Human Anaplastic Lymphoma Kinase in Complex with PF-06463922 ((10R)-7-amino-12-fluoro-2,10,16-trimethyl- 15-oxo-10,15,16,17-tetrahydro-2H-8,4-(metheno)pyrazolo(4,3-h)(2,5,11) benzoxadiazacyclotetradecine-3-carbonitrile). | | 分子名称: | (10R)-7-amino-12-fluoro-2,10,16-trimethyl-15-oxo-10,15,16,17-tetrahydro-2H-8,4-(metheno)pyrazolo[4,3-h][2,5,11]benzoxadiazacyclotetradecine-3-carbonitrile, ALK TYROSINE KINASE RECEPTOR | | 著者 | McTigue, M.A, Deng, Y.L, Liu, W, Brooun, A, Stewart, A.E. | | 登録日 | 2014-01-14 | | 公開日 | 2014-05-28 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.66 Å) | | 主引用文献 | Discovery of (10R)-7-Amino-12-Fluoro-2,10,16-Trimethyl-15-Oxo-10,15,16,17-Tetrahydro-2H-8,4-(Metheno)Pyrazolo[4,3-H][2,5,11]Benzoxadiazacyclotetradecine-3-Carbonitrile (Pf-06463922), a Macrocyclic Inhibitor of Alk/Ros1 with Pre-Clinical Brain Exposure and Broad Spectrum Potency Against Alk-Resistant Mutations.
J.Med.Chem., 57, 2014
|
|
5VRN
 
 | | CRYSTAL STRUCTURE OF THE INHA FROM MYCOBACTERIUM TUBERCULOSIS IN COMPLEX WITH AN12855, EBSI 4333. | | 分子名称: | Enoyl-[acyl-carrier-protein] reductase [NADH], [[(2~{R},3~{S},4~{R},5~{R})-5-(3-aminocarbonylpyridin-1-ium-1-yl)-4-[[5-[4-cyano-2-[(~{E})-hydroxyiminomethyl]phenoxy]-1-oxidanyl-3~{H}-2,1$l^{4}-benzoxaborol-1-yl]oxy]-3-oxidanyl-oxolan-2-yl]methoxy-oxidanyl-phosphoryl] [(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methyl hydrogen phosphate | | 著者 | Abendroth, J, Edwards, T.E, Lorimer, D. | | 登録日 | 2017-05-11 | | 公開日 | 2018-05-16 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2.55 Å) | | 主引用文献 | Discovery of a cofactor-independent inhibitor ofMycobacterium tuberculosisInhA.
Life Sci Alliance, 1, 2018
|
|
5RSA
 
 | |
