3DJE
 
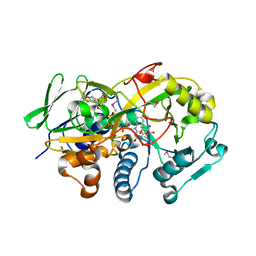 | | Crystal structure of the deglycating enzyme fructosamine oxidase from Aspergillus fumigatus (Amadoriase II) in complex with FSA | | 分子名称: | 1-S-(carboxymethyl)-1-thio-beta-D-fructopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, FLAVIN-ADENINE DINUCLEOTIDE, ... | | 著者 | Collard, F, Zhang, J, Nemet, I, Qanungo, K.R, Monnier, V.M, Yee, V.C. | | 登録日 | 2008-06-23 | | 公開日 | 2008-07-22 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Crystal structure of the deglycating enzyme fructosamine oxidase (FAOX-II)
To be Published
|
|
3P4Z
 
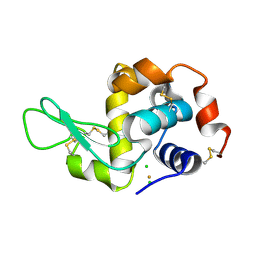 | | Time-dependent and Protein-directed In Situ Growth of Gold Nanoparticles in a Single Crystal of Lysozyme | | 分子名称: | CHLORIDE ION, GOLD 3+ ION, GOLD ION, ... | | 著者 | Wei, H, Wang, Z, Zhang, J, House, S, Gao, Y.-G, Yang, L, Robinson, H, Tan, L.H, Xing, H, Hou, C, Robertson, I.M, Zuo, J.-M, Lu, Y. | | 登録日 | 2010-10-07 | | 公開日 | 2011-02-09 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Time-dependent, protein-directed growth of gold nanoparticles within a single crystal of lysozyme.
Nat Nanotechnol, 6, 2011
|
|
3P64
 
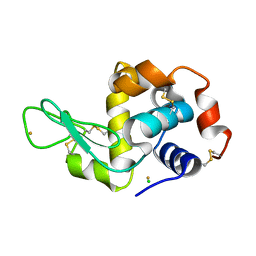 | | Time-dependent and Protein-directed In Situ Growth of Gold Nanoparticles in a Single Crystal of Lysozyme | | 分子名称: | CHLORIDE ION, GOLD 3+ ION, GOLD ION, ... | | 著者 | Wei, H, Wang, Z, Zhang, J, House, S, Gao, Y.-G, Yang, L, Robinson, H, Tan, L.H, Xing, H, Hou, C, Robertson, I.M, Zuo, J.-M, Lu, Y. | | 登録日 | 2010-10-11 | | 公開日 | 2011-02-09 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (1.3 Å) | | 主引用文献 | Time-dependent, protein-directed growth of gold nanoparticles within a single crystal of lysozyme.
Nat Nanotechnol, 6, 2011
|
|
3P68
 
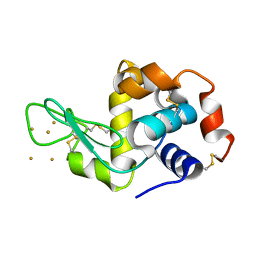 | | Time-dependent and Protein-directed In Situ Growth of Gold Nanoparticles in a Single Crystal of Lysozyme | | 分子名称: | GOLD 3+ ION, Lysozyme C | | 著者 | Wei, H, Wang, Z, Zhang, J, House, S, Gao, Y.-G, Yang, L, Robinson, H, Tan, L.H, Xing, H, Hou, C, Robertson, I.M, Zuo, J.-M, Lu, Y. | | 登録日 | 2010-10-11 | | 公開日 | 2011-02-09 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Time-dependent, protein-directed growth of gold nanoparticles within a single crystal of lysozyme.
Nat Nanotechnol, 6, 2011
|
|
3DJD
 
 | | Crystal structure of the deglycating enzyme fructosamine oxidase from Aspergillus fumigatus (Amadoriase II) | | 分子名称: | FLAVIN-ADENINE DINUCLEOTIDE, Fructosyl amine: oxygen oxidoreductase | | 著者 | Collard, F, Zhang, J, Nemet, I, Qanungo, K.R, Monnier, V.M, Yee, V.C. | | 登録日 | 2008-06-23 | | 公開日 | 2008-07-22 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Crystal structure of the deglycating enzyme fructosamine oxidase (FAOX-II)
To be Published
|
|
4F5Y
 
 | | Crystal structure of human STING CTD complex with C-di-GMP | | 分子名称: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), CALCIUM ION, Transmembrane protein 173 | | 著者 | Gu, L, Shang, G, Zhu, D, Li, N, Zhang, J, Zhu, C, Lu, D, Liu, C, Yu, Q, Zhao, Y, Xu, S. | | 登録日 | 2012-05-13 | | 公開日 | 2012-06-27 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.396 Å) | | 主引用文献 | Crystal structures of STING protein reveal basis for recognition of cyclic di-GMP
Nat.Struct.Mol.Biol., 19, 2012
|
|
4F5W
 
 | | Crystal structure of ligand free human STING CTD | | 分子名称: | CALCIUM ION, Transmembrane protein 173 | | 著者 | Gu, L, Shang, G, Zhu, D, Li, N, Zhang, J, Zhu, C, Lu, D, Liu, C, Yu, Q, Zhao, Y, Xu, S. | | 登録日 | 2012-05-13 | | 公開日 | 2012-06-27 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.201 Å) | | 主引用文献 | Crystal structures of STING protein reveal basis for recognition of cyclic di-GMP
Nat.Struct.Mol.Biol., 19, 2012
|
|
3HAG
 
 | | Crystal structure of the Hepatitis E Virus-like Particle | | 分子名称: | Capsid protein | | 著者 | Guu, T.S.Y, Liu, Z, Ye, Q, Mata, D.A, Li, K, Yin, C, Zhang, J, Tao, Y.J. | | 登録日 | 2009-05-01 | | 公開日 | 2009-09-01 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (3.5 Å) | | 主引用文献 | Structure of the hepatitis E virus-like particle suggests mechanisms for virus assembly and receptor binding.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3P65
 
 | | Time-dependent and Protein-directed In Situ Growth of Gold Nanoparticles in a Single Crystal of Lysozyme | | 分子名称: | CHLORIDE ION, GOLD 3+ ION, GOLD ION, ... | | 著者 | Wei, H, Wang, Z, Zhang, J, House, S, Gao, Y.-G, Yang, L, Robinson, H, Tan, L.H, Xing, H, Hou, C, Robertson, I.M, Zuo, J.-M, Lu, Y. | | 登録日 | 2010-10-11 | | 公開日 | 2011-02-09 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Time-dependent, protein-directed growth of gold nanoparticles within a single crystal of lysozyme.
Nat Nanotechnol, 6, 2011
|
|
7VVP
 
 | | Crystal structure of SARS-Cov-2 main protease in complex with PF07304814 | | 分子名称: | 3C-like proteinase, [(3~{S})-3-[[(2~{S})-2-[(4-methoxy-1~{H}-indol-2-yl)carbonylamino]-4-methyl-pentanoyl]amino]-2-oxidanylidene-4-[(3~{R})-2-oxidanylidene-3,4-dihydropyrrol-3-yl]butyl] dihydrogen phosphate | | 著者 | Zhou, X.L, Zhong, F.L, Lin, C, Zeng, P, Zhang, J, Li, J. | | 登録日 | 2021-11-07 | | 公開日 | 2022-08-24 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.97 Å) | | 主引用文献 | Structural Basis of Main Proteases of Coronavirus Bound to Drug Candidate PF-07304814
J.Mol.Biol., 434, 2022
|
|
8J2P
 
 | | Crystal structure of PML B-box2 | | 分子名称: | Maltose/maltodextrin-binding periplasmic protein,Protein PML, ZINC ION, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | 著者 | Zhou, C, Zang, N, Zhang, J. | | 登録日 | 2023-04-15 | | 公開日 | 2023-09-20 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.09 Å) | | 主引用文献 | Structural Basis of PML-RARA Oncoprotein Targeting by Arsenic Unravels a Cysteine Rheostat Controlling PML Body Assembly and Function.
Cancer Discov, 13, 2023
|
|
8J25
 
 | | Crystal structure of PML B-box2 mutant | | 分子名称: | Maltose/maltodextrin-binding periplasmic protein,Protein PML, ZINC ION, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | 著者 | Zhou, C, Zang, N, Zhang, J. | | 登録日 | 2023-04-14 | | 公開日 | 2023-09-20 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Structural Basis of PML-RARA Oncoprotein Targeting by Arsenic Unravels a Cysteine Rheostat Controlling PML Body Assembly and Function.
Cancer Discov, 13, 2023
|
|
2M1H
 
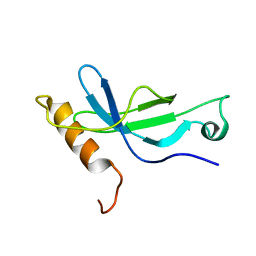 | | Solution structure of a PWWP domain from Trypanosoma brucei | | 分子名称: | Transcription elongation factor S-II | | 著者 | Wang, R, Fan, K, Liao, S, Zhang, J, Tu, X. | | 登録日 | 2012-11-28 | | 公開日 | 2013-12-04 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of TbTFIIS2-1 PWWP domain from Trypanosoma brucei.
Proteins, 84, 2016
|
|
7WQH
 
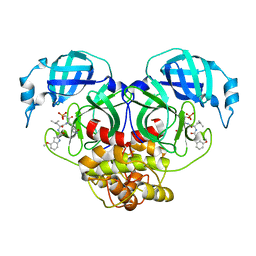 | | Crystal structure of HCoV-NL63 main protease with PF07304814 | | 分子名称: | 3C-like proteinase, [(3~{S})-3-[[(2~{S})-2-[(4-methoxy-1~{H}-indol-2-yl)carbonylamino]-4-methyl-pentanoyl]amino]-2-oxidanylidene-4-[(3~{R})-2-oxidanylidene-3,4-dihydropyrrol-3-yl]butyl] dihydrogen phosphate | | 著者 | Zhong, F.L, Zhou, X.L, Lin, C, Zeng, P, Li, J, Zhang, J. | | 登録日 | 2022-01-25 | | 公開日 | 2022-08-24 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.32 Å) | | 主引用文献 | Structural Basis of Main Proteases of Coronavirus Bound to Drug Candidate PF-07304814
J.Mol.Biol., 434, 2022
|
|
7WQJ
 
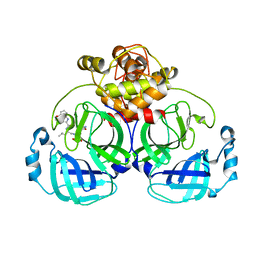 | | Crystal structure of MERS main protease in complex with PF07304814 | | 分子名称: | 3C-like proteinase, [(3~{S})-3-[[(2~{S})-2-[(4-methoxy-1~{H}-indol-2-yl)carbonylamino]-4-methyl-pentanoyl]amino]-2-oxidanylidene-4-[(3~{R})-2-oxidanylidene-3,4-dihydropyrrol-3-yl]butyl] dihydrogen phosphate | | 著者 | Lin, C, Zhang, J, Li, J. | | 登録日 | 2022-01-25 | | 公開日 | 2022-08-24 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.75 Å) | | 主引用文献 | Structural Basis of Main Proteases of Coronavirus Bound to Drug Candidate PF-07304814
J.Mol.Biol., 434, 2022
|
|
2KBE
 
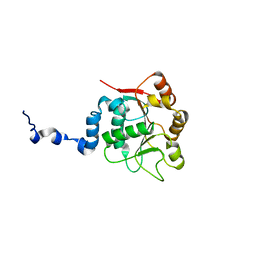 | |
2MZZ
 
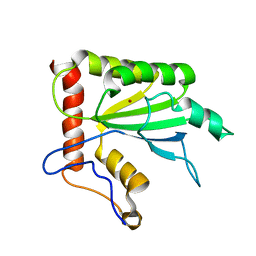 | | NMR structure of APOBEC3G NTD variant, sNTD | | 分子名称: | Apolipoprotein B mRNA-editing enzyme, catalytic polypeptide-like 3G variant, ZINC ION | | 著者 | Kouno, T, Luengas, E.M, Shigematu, M, Shandilya, S.M.D, Zhang, J, Chen, L, Hara, M, Schiffer, C.A, Harris, R.S, Matsuo, H. | | 登録日 | 2015-02-28 | | 公開日 | 2015-05-13 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structure of the Vif-binding domain of the antiviral enzyme APOBEC3G.
Nat.Struct.Mol.Biol., 22, 2015
|
|
5YMR
 
 | | The Crystal Structure of IseG | | 分子名称: | 2-hydroxyethylsulfonic acid, Formate acetyltransferase, GLYCEROL | | 著者 | Lin, L, Zhang, J, Xing, M, Hua, G, Guo, C, Hu, Y, Wei, Y, Ang, E, Zhao, H, Zhang, Y, Yuchi, Z. | | 登録日 | 2017-10-22 | | 公開日 | 2019-03-20 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Radical-mediated C-S bond cleavage in C2 sulfonate degradation by anaerobic bacteria.
Nat Commun, 10, 2019
|
|
6C5W
 
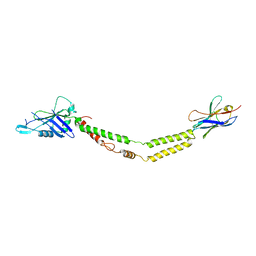 | | Crystal structure of the mitochondrial calcium uniporter | | 分子名称: | CALCIUM ION, calcium uniporter, nanobody | | 著者 | Fan, C, Fan, M, Fastman, N, Zhang, J, Feng, L. | | 登録日 | 2018-01-17 | | 公開日 | 2018-07-11 | | 最終更新日 | 2019-04-24 | | 実験手法 | X-RAY DIFFRACTION (3.10010242 Å) | | 主引用文献 | X-ray and cryo-EM structures of the mitochondrial calcium uniporter.
Nature, 559, 2018
|
|
6K7V
 
 | | Structure of NLRP1 CARD filament | | 分子名称: | NACHT, LRR and PYD domains-containing protein 1 | | 著者 | Gong, Q, Xu, C, Zhang, J, Wu, B. | | 登録日 | 2019-06-09 | | 公開日 | 2020-09-16 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (3.7 Å) | | 主引用文献 | Structural basis for distinct inflammasome complex assembly by human NLRP1 and CARD8.
Nat Commun, 12, 2021
|
|
7XAX
 
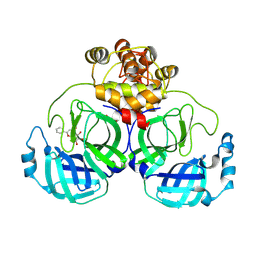 | |
7XB3
 
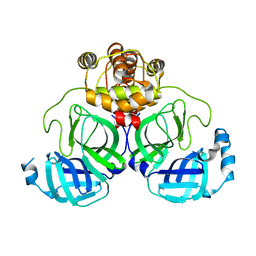 | |
2NAS
 
 | |
7XB4
 
 | | Crystal structure of SARS-Cov-2 main protease D48N mutant in complex with PF07321332 | | 分子名称: | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, Replicase polyprotein 1a | | 著者 | Hu, X.H, Li, J, Zhang, J. | | 登録日 | 2022-03-20 | | 公開日 | 2023-03-22 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.07 Å) | | 主引用文献 | Crystal structure of SARS-Cov-2 main protease D48N mutant in complex with PF07321332
To Be Published
|
|
2KBF
 
 | |
