6BBE
 
 | | Structure of N-glycosylated porcine surfactant protein-D | | 分子名称: | 2-(2-{2-[2-(2-METHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHANOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | 著者 | van Eijk, M, Rynkiewicz, M.J, Khatri, K, Leymarie, N, Zaia, J, White, M.R, Hartshorn, K.L, Cafarella, T.R, van Die, I, Hessing, M, Seaton, B.A, Haagsman, H.P. | | 登録日 | 2017-10-18 | | 公開日 | 2018-05-23 | | 最終更新日 | 2021-03-24 | | 実験手法 | X-RAY DIFFRACTION (1.898 Å) | | 主引用文献 | Lectin-mediated binding and sialoglycans of porcine surfactant protein D synergistically neutralize influenza A virus.
J. Biol. Chem., 293, 2018
|
|
5U07
 
 | | CRISPR RNA-guided surveillance complex | | 分子名称: | CRISPR-associated protein, Cas5e family, Cse1 family, ... | | 著者 | Xiao, Y, Luo, M, Hayes, R.P, Kim, J, Ng, S, Ding, F, Liao, M, Ke, A. | | 登録日 | 2016-11-23 | | 公開日 | 2017-08-09 | | 最終更新日 | 2024-03-13 | | 実験手法 | ELECTRON MICROSCOPY (3.8 Å) | | 主引用文献 | Structure Basis for Directional R-loop Formation and Substrate Handover Mechanisms in Type I CRISPR-Cas System.
Cell, 170, 2017
|
|
5UGK
 
 | | Zinc-Binding Structure of a Catalytic Amyloid from Solid-State NMR Spectroscopy | | 分子名称: | ILE-HIS-VAL-HIS-LEU-GLN-ILE, ZINC ION | | 著者 | Lee, M, Wang, T, Makhlynets, O.V, Wu, Y, Polizzi, N, Wu, H, Gosavi, P.M, Korendovych, I.V, DeGrado, W.F, Hong, M. | | 登録日 | 2017-01-09 | | 公開日 | 2017-05-31 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLID-STATE NMR | | 主引用文献 | Zinc-binding structure of a catalytic amyloid from solid-state NMR.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
6RMK
 
 | | Bacteriorhodopsin, dark state, cell 2, refined using the same protocol as sub-ps time delays | | 分子名称: | Bacteriorhodopsin, RETINAL | | 著者 | Nass Kovacs, G, Colletier, J.-P, Gruenbein, M.L, Stensitzki, T, Batyuk, A, Carbajo, S, Doak, R.B, Ehrenberg, D, Foucar, L, Gasper, R, Gorel, A, Hilpert, M, Kloos, M, Koglin, J, Reinstein, J, Roome, C.M, Schlesinger, R, Seaberg, M, Shoeman, R.L, Stricker, M, Boutet, S, Haacke, S, Heberle, J, Domratcheva, T, Barends, T.R.M, Schlichting, I. | | 登録日 | 2019-05-07 | | 公開日 | 2019-06-05 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Three-dimensional view of ultrafast dynamics in photoexcited bacteriorhodopsin.
Nat Commun, 10, 2019
|
|
8HJ9
 
 | | cryoEM structure of glutamate dehydrogenase from Thermococcus profundus in complex with NADP | | 分子名称: | Glutamate dehydrogenase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | 著者 | Wakabayashi, T, Oide, M, Kato, T, Nakasako, M. | | 登録日 | 2022-11-22 | | 公開日 | 2023-02-08 | | 最終更新日 | 2023-12-20 | | 実験手法 | ELECTRON MICROSCOPY (3.12 Å) | | 主引用文献 | Coenzyme-binding pathway on glutamate dehydrogenase suggested from multiple-binding sites visualized by cryo-electron microscopy.
Febs J., 290, 2023
|
|
8HJ3
 
 | | cryoEM structure of glutamate dehydrogenase from Thermococcus profundus in complex with NADP | | 分子名称: | Glutamate dehydrogenase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | 著者 | Wakabayashi, T, Oide, M, Kato, T, Nakasako, M. | | 登録日 | 2022-11-22 | | 公開日 | 2023-02-08 | | 最終更新日 | 2023-12-20 | | 実験手法 | ELECTRON MICROSCOPY (3.29 Å) | | 主引用文献 | Coenzyme-binding pathway on glutamate dehydrogenase suggested from multiple-binding sites visualized by cryo-electron microscopy.
Febs J., 290, 2023
|
|
8HHO
 
 | | cryoEM structure of glutamate dehydrogenase from Thermococcus profundus in complex with NADP | | 分子名称: | Glutamate dehydrogenase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | 著者 | Wakabayashi, T, Oide, M, Kato, T, Nakasako, M. | | 登録日 | 2022-11-16 | | 公開日 | 2023-02-08 | | 最終更新日 | 2023-12-20 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Coenzyme-binding pathway on glutamate dehydrogenase suggested from multiple-binding sites visualized by cryo-electron microscopy.
Febs J., 290, 2023
|
|
3E3U
 
 | | Crystal structure of Mycobacterium tuberculosis peptide deformylase in complex with inhibitor | | 分子名称: | N-[(2R)-2-{[(2S)-2-(1,3-benzoxazol-2-yl)pyrrolidin-1-yl]carbonyl}hexyl]-N-hydroxyformamide, NICKEL (II) ION, Peptide deformylase | | 著者 | Meng, W, Xu, M, Pan, S, Koehn, J. | | 登録日 | 2008-08-08 | | 公開日 | 2009-01-20 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (1.56 Å) | | 主引用文献 | Peptide deformylase inhibitors of Mycobacterium tuberculosis: synthesis, structural investigations, and biological results.
Bioorg.Med.Chem.Lett., 18, 2008
|
|
8HIQ
 
 | | cryoEM structure of glutamate dehydrogenase from Thermococcus profundus in complex with NADP | | 分子名称: | Glutamate dehydrogenase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | 著者 | Wakabayashi, T, Oide, M, Kato, T, Nakasako, M. | | 登録日 | 2022-11-21 | | 公開日 | 2023-02-08 | | 最終更新日 | 2023-12-20 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Coenzyme-binding pathway on glutamate dehydrogenase suggested from multiple-binding sites visualized by cryo-electron microscopy.
Febs J., 290, 2023
|
|
8HIZ
 
 | | cryoEM structure of glutamate dehydrogenase from Thermococcus profundus in complex with NADP | | 分子名称: | Glutamate dehydrogenase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | 著者 | Wakabayashi, T, Oide, M, Kato, T, Nakasako, M. | | 登録日 | 2022-11-22 | | 公開日 | 2023-02-08 | | 最終更新日 | 2023-12-20 | | 実験手法 | ELECTRON MICROSCOPY (3.08 Å) | | 主引用文献 | Coenzyme-binding pathway on glutamate dehydrogenase suggested from multiple-binding sites visualized by cryo-electron microscopy.
Febs J., 290, 2023
|
|
5MMQ
 
 | | ABA RECEPTOR FROM CITRUS, CSPYL1 | | 分子名称: | CSPYL1 | | 著者 | Moreno-Alvero, M, Yunta, C, Gonzalez-Guzman, M, Arbona, V, Granell, A, Martinez-Ripoll, M, Infantes, L, Rodriguez, P.L. | | 登録日 | 2016-12-12 | | 公開日 | 2017-08-02 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Structure of Ligand-Bound Intermediates of Crop ABA Receptors Highlights PP2C as Necessary ABA Co-receptor.
Mol Plant, 10, 2017
|
|
5MN0
 
 | | ABA RECEPTOR FROM CITRUS, CSPYL1 | | 分子名称: | (2Z,4E)-5-[(1S)-1-hydroxy-2,6,6-trimethyl-4-oxocyclohex-2-en-1-yl]-3-methylpenta-2,4-dienoic acid, CHLORIDE ION, CSPYL1, ... | | 著者 | Moreno-Alvero, M, Yunta, C, Gonzalez-Guzman, M, Arbona, V, Granell, A, Martinez-Ripoll, M, Infantes, L, Rodriguez, P.L, Albert, A. | | 登録日 | 2016-12-12 | | 公開日 | 2017-08-02 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structure of Ligand-Bound Intermediates of Crop ABA Receptors Highlights PP2C as Necessary ABA Co-receptor.
Mol Plant, 10, 2017
|
|
6S60
 
 | | Crystal structure of hTEAD2 in complex with a trisubstituted pyrazole inhibitor | | 分子名称: | 4-[3-(3,4-dichlorophenyl)-4-[(phenylmethyl)carbamoyl]pyrazol-1-yl]butanoic acid, MYRISTIC ACID, Transcriptional enhancer factor TEF-4 | | 著者 | Sturbaut, M, Allemand, F, Guichou, J.F. | | 登録日 | 2019-07-02 | | 公開日 | 2020-07-22 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Discovery of a cryptic site at the interface 2 of TEAD - Towards a new family of YAP/TAZ-TEAD inhibitors.
Eur.J.Med.Chem., 226, 2021
|
|
6P73
 
 | | Cytochrome-C-nitrite reductase | | 分子名称: | CALCIUM ION, Cytochrome c-552, HEME C | | 著者 | Schmidt, M, Pacheco, A. | | 登録日 | 2019-06-04 | | 公開日 | 2020-04-01 | | 最終更新日 | 2024-10-23 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | Trapping of a Putative Intermediate in the CytochromecNitrite Reductase (ccNiR)-Catalyzed Reduction of Nitrite: Implications for the ccNiR Reaction Mechanism.
J.Am.Chem.Soc., 141, 2019
|
|
6MRY
 
 | | NoD173 plant defensin | | 分子名称: | 1,2-ETHANEDIOL, 5-amino-2,4,6-triiodobenzene-1,3-dicarboxylic acid, CHLORIDE ION, ... | | 著者 | Caria, S, Kvansakul, M. | | 登録日 | 2018-10-15 | | 公開日 | 2019-04-24 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Structural and functional characterization of the membrane-permeabilizing activity ofNicotiana occidentalisdefensin NoD173 and protein engineering to enhance oncolysis.
Faseb J., 33, 2019
|
|
5CEH
 
 | | Structure of histone lysine demethylase KDM5A in complex with selective inhibitor | | 分子名称: | 7-oxo-5-phenyl-6-(propan-2-yl)-4,7-dihydropyrazolo[1,5-a]pyrimidine-3-carbonitrile, Lysine-specific demethylase 5A, NICKEL (II) ION, ... | | 著者 | Kiefer, J.R, Vinogradova, M. | | 登録日 | 2015-07-06 | | 公開日 | 2016-05-18 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (3.14 Å) | | 主引用文献 | An inhibitor of KDM5 demethylases reduces survival of drug-tolerant cancer cells.
Nat.Chem.Biol., 12, 2016
|
|
7Z8O
 
 | | Crystal structure of SARS-CoV-2 S RBD in complex with a stapled peptide | | 分子名称: | 2,4,6-tris(chloromethyl)-1,3,5-triazine, GLYCEROL, Spike protein S1, ... | | 著者 | Brear, P, Chen, L, Gaynor, K, Harman, M, Dods, R, Hyvonen, M. | | 登録日 | 2022-03-18 | | 公開日 | 2023-06-28 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (0.96 Å) | | 主引用文献 | Multivalent bicyclic peptides are an effective antiviral modality that can potently inhibit SARS-CoV-2.
Nat Commun, 14, 2023
|
|
6PRX
 
 | | oxidized Human Branched Chain Aminotransferase mutant C318A | | 分子名称: | Branched-chain-amino-acid aminotransferase, mitochondrial, PYRIDOXAL-5'-PHOSPHATE | | 著者 | Dong, M, Herbert, D, Gibbs, S. | | 登録日 | 2019-07-11 | | 公開日 | 2020-01-22 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (3.25 Å) | | 主引用文献 | Crystal structure of an oxidized mutant of human mitochondrial branched-chain aminotransferase.
Acta Crystallogr.,Sect.F, 76, 2020
|
|
8B1X
 
 | | Solution NMR structure of the single alpha helix peptide (P3-7)2 | | 分子名称: | P3-7_2 | | 著者 | Escobedo, A, Coles, M, Diercks, T, Garcia, J, Millet, O, Salvatella, X. | | 登録日 | 2022-09-12 | | 公開日 | 2023-01-25 | | 最終更新日 | 2024-06-19 | | 実験手法 | SOLUTION NMR | | 主引用文献 | A glutamine-based single alpha-helix scaffold to target globular proteins.
Nat Commun, 13, 2022
|
|
6P9G
 
 | | Structure of USP5 zinc-finger ubiquitin binding domain co-crystallized with 2-(4-oxoquinazolin-3(4H)-yl)propanoic acid | | 分子名称: | (2R)-2-(4-oxoquinazolin-3(4H)-yl)propanoic acid, UNKNOWN ATOM OR ION, Ubiquitin carboxyl-terminal hydrolase 5, ... | | 著者 | Tempel, W, Mann, M.K, Harding, R.J, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Schapira, M, Structural Genomics Consortium (SGC) | | 登録日 | 2019-06-10 | | 公開日 | 2019-09-18 | | 最終更新日 | 2024-10-23 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Discovery of Small Molecule Antagonists of the USP5 Zinc Finger Ubiquitin-Binding Domain.
J.Med.Chem., 62, 2019
|
|
8P2K
 
 | | Ternary complex of translating ribosome, NAC and METAP1 | | 分子名称: | 18s rRNA, 28S rRNA, 40S ribosomal protein S11, ... | | 著者 | Jia, M, Jaskolowski, M, Scaiola, A, Jomaa, A, Ban, N. | | 登録日 | 2023-05-16 | | 公開日 | 2023-07-19 | | 最終更新日 | 2024-04-24 | | 実験手法 | ELECTRON MICROSCOPY (2.9 Å) | | 主引用文献 | NAC controls cotranslational N-terminal methionine excision in eukaryotes.
Science, 380, 2023
|
|
5Y0D
 
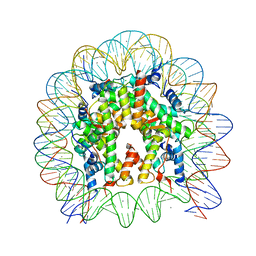 | | Crystal Structure of the human nucleosome containing the H2B E76K mutant | | 分子名称: | CHLORIDE ION, DNA (146-MER), Histone H2A type 1-B/E, ... | | 著者 | Kurumizaka, H, Arimura, Y, Fujita, R, Noda, M. | | 登録日 | 2017-07-16 | | 公開日 | 2018-07-18 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.99 Å) | | 主引用文献 | Cancer-associated mutations of histones H2B, H3.1 and H2A.Z.1 affect the structure and stability of the nucleosome.
Nucleic Acids Res., 46, 2018
|
|
7APF
 
 | | Crystal structure of JAK3 in complex with FM601 (compound 10a) | | 分子名称: | 1,2-ETHANEDIOL, 1-phenylurea, 3-[3-(propanoylamino)phenyl]-1~{H}-pyrrolo[2,3-b]pyridine-5-carboxamide, ... | | 著者 | Chaikuad, A, Forster, M, Gehringer, M, Laufer, S, Knapp, S, Structural Genomics Consortium (SGC) | | 登録日 | 2020-10-16 | | 公開日 | 2020-12-02 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Discovery of a Novel Class of Covalent Dual Inhibitors Targeting the Protein Kinases BMX and BTK.
Int J Mol Sci, 21, 2020
|
|
8OWN
 
 | |
3E95
 
 | | Crystal Structure of the Plasmodium Falciparum ubiquitin conjugating enzyme complex, PfUBC13-PfUev1a | | 分子名称: | UNKNOWN ATOM OR ION, Ubiquitin carrier protein, Ubiquitin-conjugating enzyme E2 | | 著者 | Wernimont, A.K, Lam, A, Ali, A, Brokx, S, Lin, Y.H, Zhao, Y, Lew, J, Ravichandran, M, Wasney, G, Vedadi, M, Kozieradzki, I, Schapira, M, Bochkarev, A, Wilkstrom, M, BOuntra, C, Arrowsmith, C.H, Edwards, A.M, Hui, R, Qiu, W, Brand, V.B, Structural Genomics Consortium (SGC) | | 登録日 | 2008-08-21 | | 公開日 | 2008-09-30 | | 最終更新日 | 2024-10-30 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Crystal Structure of the Plasmodium Falciparum ubiquitin conjugating enzyme complex, PfUBC13-PfUev1a
TO BE PUBLISHED
|
|
