7JMG
 
 | | Functional Pathways of Biomolecules Retrieved from Single-particle Snapshots - Frame 22 - State 2 (S2) | | 分子名称: | CALCIUM ION, Peptidyl-prolyl cis-trans isomerase FKBP1B, ZINC ION, ... | | 著者 | Dashti, A, des Georges, A, Frank, J, Ourmazd, A. | | 登録日 | 2020-07-31 | | 公開日 | 2020-08-12 | | 最終更新日 | 2024-03-06 | | 実験手法 | ELECTRON MICROSCOPY (4.5 Å) | | 主引用文献 | Retrieving functional pathways of biomolecules from single-particle snapshots.
Nat Commun, 11, 2020
|
|
7PSZ
 
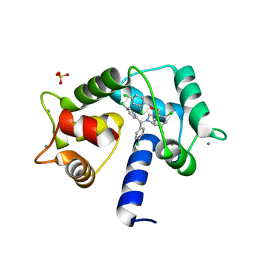 | | Crystal structure of CaM in complex with CDZ (form 1) | | 分子名称: | 1-[bis(4-chlorophenyl)methyl]-3-[(2~{R})-2-(2,4-dichlorophenyl)-2-[(2,4-dichlorophenyl)methoxy]ethyl]imidazole, CALCIUM ION, Calmodulin-1, ... | | 著者 | Mechaly, A.E, Leger, C, Haouz, A, Chenal, A. | | 登録日 | 2021-09-24 | | 公開日 | 2022-08-17 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.898 Å) | | 主引用文献 | Dynamics and structural changes of calmodulin upon interaction with the antagonist calmidazolium.
Bmc Biol., 20, 2022
|
|
7PU9
 
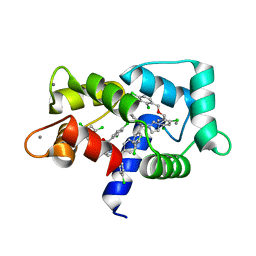 | | Crystal structure of CaM in complex with CDZ (form 2) | | 分子名称: | 1-[bis(4-chlorophenyl)methyl]-3-[(2~{R})-2-(2,4-dichlorophenyl)-2-[(2,4-dichlorophenyl)methoxy]ethyl]imidazole, CALCIUM ION, Calmodulin-1 | | 著者 | Mechaly, A.E, Leger, C, Haouz, A, Chenal, A. | | 登録日 | 2021-09-28 | | 公開日 | 2022-08-17 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2.279 Å) | | 主引用文献 | Dynamics and structural changes of calmodulin upon interaction with the antagonist calmidazolium.
Bmc Biol., 20, 2022
|
|
7US2
 
 | | PARL-cleaved Skd3 (human ClpB) E455Q Nucleotide Binding Domain hexamer bound to ATPgammaS, open conformation | | 分子名称: | Caseinolytic peptidase B protein homolog, MAGNESIUM ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, ... | | 著者 | Gupta, A, Lentzsch, A.M, Siegel, A.S, Yu, Z, Lu, C, Chio, U.S, Cheng, Y, Shan, S.-o. | | 登録日 | 2022-04-22 | | 公開日 | 2023-04-26 | | 最終更新日 | 2023-11-08 | | 実験手法 | ELECTRON MICROSCOPY (2.76 Å) | | 主引用文献 | Dodecamer assembly of a metazoan AAA + chaperone couples substrate extraction to refolding.
Sci Adv, 9, 2023
|
|
6SR5
 
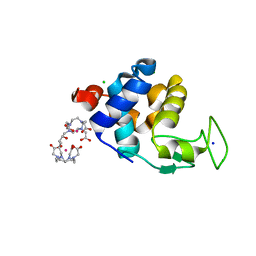 | | X-ray pump X-ray probe on lysozyme.Gd nanocrystals: 102 fs time delay | | 分子名称: | 10-((2R)-2-HYDROXYPROPYL)-1,4,7,10-TETRAAZACYCLODODECANE 1,4,7-TRIACETIC ACID, CHLORIDE ION, GADOLINIUM ATOM, ... | | 著者 | Kloos, M, Gorel, A, Nass, K. | | 登録日 | 2019-09-04 | | 公開日 | 2020-04-22 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Structural dynamics in proteins induced by and probed with X-ray free-electron laser pulses.
Nat Commun, 11, 2020
|
|
4M1E
 
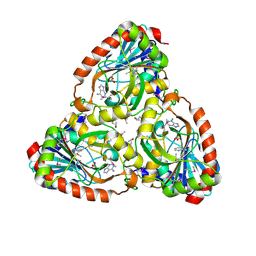 | | Crystal structure of purine nucleoside phosphorylase I from Planctomyces limnophilus DSM 3776, NYSGRC Target 029364. | | 分子名称: | ADENINE, PYRIDINE-2-CARBOXYLIC ACID, Purine nucleoside phosphorylase, ... | | 著者 | Malashkevich, V.N, Bonanno, J.B, Bhosle, R, Toro, R, Hillerich, B, Gizzi, A, Garforth, S, Kar, A, Chan, M.K, Lafluer, J, Patel, H, Matikainen, B, Chamala, S, Lim, S, Celikgil, A, Villegas, G, Evans, B, Love, J, Fiser, A, Khafizov, K, Seidel, R, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | 登録日 | 2013-08-02 | | 公開日 | 2013-08-21 | | 最終更新日 | 2023-12-06 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Crystal structure of purine nucleoside phosphorylase I from Planctomyces limnophilus DSM 3776, NYSGRC Target 029364.
To be Published
|
|
6ZQP
 
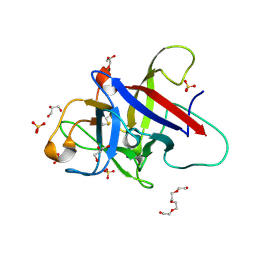 | | Structure of the Pmt2-MIR domain with bound ligands | | 分子名称: | GLYCEROL, PMT2 isoform 1, SULFATE ION, ... | | 著者 | Wild, K, Chiapparino, A, Hackmann, Y, Mortensen, S, Sinning, I. | | 登録日 | 2020-07-10 | | 公開日 | 2020-12-23 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Functional implications of MIR domains in protein O -mannosylation.
Elife, 9, 2020
|
|
4MAR
 
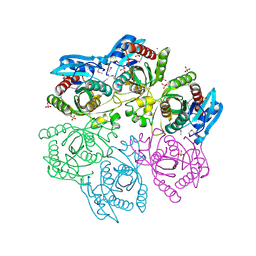 | | Crystal structure of purine nucleoside phosphorylase from Meiothermus ruber DSM 1279 complexed with sulfate. | | 分子名称: | MAGNESIUM ION, Purine nucleoside phosphorylase DeoD-type, SULFATE ION | | 著者 | Malashkevich, V.N, Bhosle, R, Toro, R, Hillerich, B, Gizzi, A, Garforth, S, Kar, A, Chan, M.K, Lafluer, J, Patel, H, Matikainen, B, Chamala, S, Lim, S, Celikgil, A, Villegas, G, Evans, B, Love, J, Fiser, A, Khafizov, K, Seidel, R, Bonanno, J.B, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | 登録日 | 2013-08-16 | | 公開日 | 2013-10-09 | | 最終更新日 | 2023-12-06 | | 実験手法 | X-RAY DIFFRACTION (2.16 Å) | | 主引用文献 | Crystal structure of purine nucleoside phosphorylase from Meiothermus ruber DSM 1279 complexed with sulfate.
To be Published
|
|
6YTC
 
 | |
6ZQQ
 
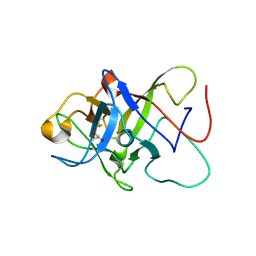 | | Structure of the Pmt3-MIR domain with bound ligands | | 分子名称: | GLYCEROL, PMT3 isoform 1 | | 著者 | Wild, K, Chiapparino, A, Hackmann, Y, Mortensen, S, Sinning, I. | | 登録日 | 2020-07-10 | | 公開日 | 2020-12-23 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Functional implications of MIR domains in protein O -mannosylation.
Elife, 9, 2020
|
|
6ZM6
 
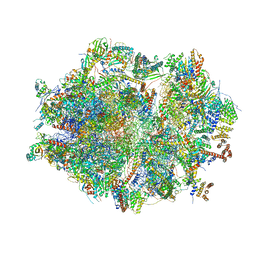 | | Human mitochondrial ribosome in complex with mRNA, A/A tRNA and P/P tRNA | | 分子名称: | 12S mitochondrial rRNA, 16S mitochondrial rRNA, 28S ribosomal protein S10, ... | | 著者 | Itoh, Y, Andrell, J, Amunts, A. | | 登録日 | 2020-07-01 | | 公開日 | 2021-01-13 | | 最終更新日 | 2023-11-15 | | 実験手法 | ELECTRON MICROSCOPY (2.59 Å) | | 主引用文献 | Mechanism of membrane-tethered mitochondrial protein synthesis.
Science, 371, 2021
|
|
4MBA
 
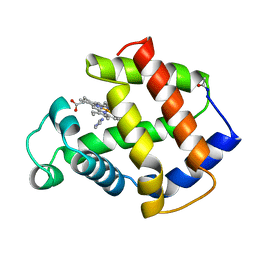 | | APLYSIA LIMACINA MYOGLOBIN. CRYSTALLOGRAPHIC ANALYSIS AT 1.6 ANGSTROMS RESOLUTION | | 分子名称: | IMIDAZOLE, MYOGLOBIN, PROTOPORPHYRIN IX CONTAINING FE | | 著者 | Bolognesi, M, Onesti, S, Gatti, G, Coda, A, Ascenzi, P, Brunori, M. | | 登録日 | 1989-02-22 | | 公開日 | 1990-01-15 | | 最終更新日 | 2024-06-05 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Aplysia limacina myoglobin. Crystallographic analysis at 1.6 A resolution.
J.Mol.Biol., 205, 1989
|
|
1BOZ
 
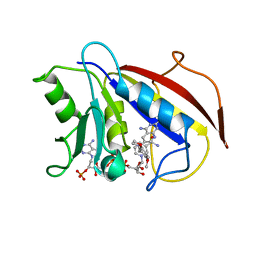 | | STRUCTURE-BASED DESIGN AND SYNTHESIS OF LIPOPHILIC 2,4-DIAMINO-6-SUBSTITUTED QUINAZOLINES AND THEIR EVALUATION AS INHIBITORS OF DIHYDROFOLATE REDUCTASE AND POTENTIAL ANTITUMOR AGENTS | | 分子名称: | N6-(2,5-DIMETHOXY-BENZYL)-N6-METHYL-PYRIDO[2,3-D]PYRIMIDINE-2,4,6-TRIAMINE, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, PROTEIN (DIHYDROFOLATE REDUCTASE) | | 著者 | Gangjee, A, Vidwans, A.P, Vasudevan, A, Queener, S.F, Kisliuk, R.L, Cody, V, Li, R, Galitsky, N, Luft, J.R, Pangborn, W. | | 登録日 | 1998-08-06 | | 公開日 | 1998-08-12 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Structure-based design and synthesis of lipophilic 2,4-diamino-6-substituted quinazolines and their evaluation as inhibitors of dihydrofolate reductases and potential antitumor agents.
J.Med.Chem., 41, 1998
|
|
7SKM
 
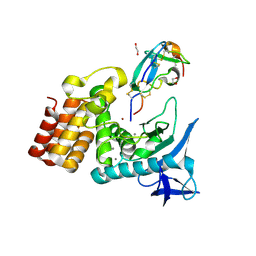 | | Complex between S. aureus aureolysin and wt IMPI. | | 分子名称: | 1,2-ETHANEDIOL, CALCIUM ION, DI(HYDROXYETHYL)ETHER, ... | | 著者 | Mendes, S.R, Eckhard, U, Rodriguez-Banqueri, A, Guevara, T, Gomis-Ruth, F.X. | | 登録日 | 2021-10-21 | | 公開日 | 2022-01-26 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | An engineered protein-based submicromolar competitive inhibitor of the Staphylococcus aureus virulence factor aureolysin
Comput Struct Biotechnol J, 20, 2022
|
|
7QDS
 
 | | Apo human SKI complex in the open state | | 分子名称: | Helicase SKI2W, Tetratricopeptide repeat protein 37, WD repeat-containing protein 61 | | 著者 | Koegel, A, Keidel, A, Bonneau, A, Schaefer, I.B, Conti, E. | | 登録日 | 2021-11-30 | | 公開日 | 2022-02-02 | | 最終更新日 | 2022-03-02 | | 実験手法 | ELECTRON MICROSCOPY (3.8 Å) | | 主引用文献 | The human SKI complex regulates channeling of ribosome-bound RNA to the exosome via an intrinsic gatekeeping mechanism.
Mol.Cell, 82, 2022
|
|
8V5Y
 
 | |
6R88
 
 | | Structure of Arabidopsis thaliana GLR3.3 ligand-binding domain in complex with glycine | | 分子名称: | CHLORIDE ION, GLYCEROL, GLYCINE, ... | | 著者 | Alfieri, A, Pederzoli, R, Costa, A. | | 登録日 | 2019-04-01 | | 公開日 | 2020-01-01 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | The structural bases for agonist diversity in anArabidopsis thalianaglutamate receptor-like channel.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
7SKL
 
 | | Complex between S. aureus aureolysin and IMPI mutant I57I | | 分子名称: | 1,2-ETHANEDIOL, CALCIUM ION, IMPI alpha, ... | | 著者 | Mendes, S.R, Eckhard, U, Rodriguez-Banqueri, A, Guevara, T, Gomis-Ruth, F.X. | | 登録日 | 2021-10-21 | | 公開日 | 2022-01-26 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | An engineered protein-based submicromolar competitive inhibitor of the Staphylococcus aureus virulence factor aureolysin
Comput Struct Biotechnol J, 20, 2022
|
|
3ZWE
 
 | | Structure of BambL, a lectin from Burkholderia ambifaria, complexed with blood group B epitope | | 分子名称: | BAMBL LECTIN, alpha-L-fucopyranose, alpha-L-fucopyranose-(1-2)-[alpha-D-galactopyranose-(1-3)]beta-D-galactopyranose-(1-4)-beta-D-glucopyranose, ... | | 著者 | Audfray, A, Claudinon, J, Abounit, S, Ruvoen-Clouet, N, Larson, G, Wimmerova, M, LePendu, J, Romer, W, Varrot, A, Imberty, A. | | 登録日 | 2011-07-29 | | 公開日 | 2011-12-21 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Fucose-Binding Lectin from Opportunistic Pathogen Burkholderia Ambifaria Binds to Both Plant and Human Oligosaccharidic Epitopes.
J.Biol.Chem., 287, 2012
|
|
1Z6G
 
 | | Crystal structure of guanylate kinase from Plasmodium falciparum | | 分子名称: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, SULFATE ION, guanylate kinase | | 著者 | Mulichak, A.M, Lew, J, Artz, J, Choe, J, Walker, J.R, Zhao, Y, Sundstrom, M, Arrowsmith, C, Edwards, A, Bochkarev, A, Hui, R, Gao, M, Structural Genomics Consortium (SGC) | | 登録日 | 2005-03-22 | | 公開日 | 2005-04-05 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (2.18 Å) | | 主引用文献 | Genome-scale protein expression and structural biology of Plasmodium falciparum and related Apicomplexan organisms.
Mol.Biochem.Parasitol., 151, 2007
|
|
3ZW2
 
 | | Structure of the lectin Bambl from Burkholderia ambifaria in complex with blood group H type 1 tetrasaccharide | | 分子名称: | BAMBL LECTIN, alpha-L-fucopyranose-(1-2)-beta-D-galactopyranose, alpha-L-fucopyranose-(1-2)-beta-D-galactopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Audfray, A, Claudinon, J, Abounit, S, Ruvoen-Clouet, N, Larson, G, Wimmerova, M, Lependu, J, Romer, W, Varrot, A, Imberty, A. | | 登録日 | 2011-07-28 | | 公開日 | 2011-11-16 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Fucose-Binding Lectin from Opportunistic Pathogen Burkholderia Ambifaria Binds to Both Plant and Human Oligosaccharidic Epitopes.
J.Biol.Chem., 287, 2012
|
|
3ZW0
 
 | | Structure of BambL lectin from Burkholderia ambifaria | | 分子名称: | BAMBL LECTIN, alpha-L-fucopyranose | | 著者 | Audfray, A, Claudinon, J, Abounit, S, Ruvoen-Clouet, N, Larson, G, Wimmerova, M, LePendu, J, Romer, W, Varrot, A, Imberty, A. | | 登録日 | 2011-07-28 | | 公開日 | 2011-12-21 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Fucose-Binding Lectin from Opportunistic Pathogen Burkholderia Ambifaria Binds to Both Plant and Human Oligosaccharidic Epitopes.
J.Biol.Chem., 287, 2012
|
|
4LN1
 
 | | CRYSTAL STRUCTURE OF L-lactate dehydrogenase from Bacillus cereus ATCC 14579 complexed with calcium, NYSGRC Target 029452 | | 分子名称: | CALCIUM ION, L-lactate dehydrogenase 1 | | 著者 | Malashkevich, V.N, Bonanno, J.B, Bhosle, R, Toro, R, Hillerich, B, Gizzi, A, Garforth, S, Kar, A, Chan, M.K, Lafluer, J, Patel, H, Matikainen, B, Chamala, S, Lim, S, Celikgil, A, Villegas, G, Evans, B, Love, J, Fiser, A, Khafizov, K, Seidel, R, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | 登録日 | 2013-07-11 | | 公開日 | 2013-07-24 | | 最終更新日 | 2023-12-06 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Crystal structure of L-lactate dehydrogenase from Bacillus cereus ATCC 14579 complexed with calcium, NYSGRC Target 029452
To be Published
|
|
7SNU
 
 | | Crystal structure of ShHTL7 from Striga hermonthica in complex with strigolactone antagonist RG6 | | 分子名称: | 2-{(2S)-1-[(4-ethoxyphenyl)methyl]-4-[(2E)-3-(4-methoxyphenyl)prop-2-en-1-yl]piperazin-2-yl}ethan-1-ol, ACETATE ION, GLYCEROL, ... | | 著者 | Arellano-Saab, A, Stogios, P.J, Skarina, T, Yim, V, Savchenko, A, McCourt, P. | | 登録日 | 2021-10-28 | | 公開日 | 2022-07-06 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.46 Å) | | 主引用文献 | A novel strigolactone receptor antagonist provides insights into the structural inhibition, conditioning, and germination of the crop parasite Striga.
J.Biol.Chem., 298, 2022
|
|
8V8E
 
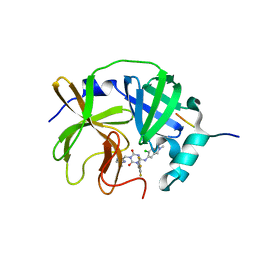 | | Room-temperature X-ray structure of SARS-CoV-2 main protease catalytic domain (residues 1-199-6H) in complex with ensitrelvir (ESV) | | 分子名称: | 3C-like proteinase nsp5, 6-[(6-chloranyl-2-methyl-indazol-5-yl)amino]-3-[(1-methyl-1,2,4-triazol-3-yl)methyl]-1-[[2,4,5-tris(fluoranyl)phenyl]methyl]-1,3,5-triazine-2,4-dione, peptide | | 著者 | Kovalevsky, A, Coates, L. | | 登録日 | 2023-12-05 | | 公開日 | 2024-05-29 | | 最終更新日 | 2024-06-12 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Visualizing the Active Site Oxyanion Loop Transition Upon Ensitrelvir Binding and Transient Dimerization of SARS-CoV-2 Main Protease.
J.Mol.Biol., 436, 2024
|
|
