8X5D
 
 | |
5ZUG
 
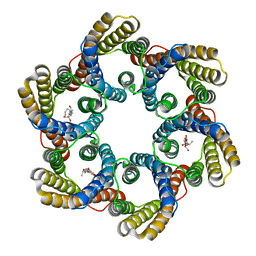 | | Structure of the bacterial acetate channel SatP | | 分子名称: | Succinate-acetate/proton symporter SatP, nonyl beta-D-glucopyranoside | | 著者 | Sun, P.C, Li, J.L, Xiao, Q.J, Guan, Z.Y, Deng, D. | | 登録日 | 2018-05-07 | | 公開日 | 2018-11-21 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (2.802 Å) | | 主引用文献 | Crystal structure of the bacterial acetate transporter SatP reveals that it forms a hexameric channel.
J. Biol. Chem., 293, 2018
|
|
4QXE
 
 | |
6BE0
 
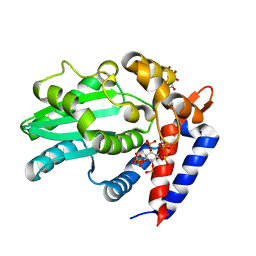 | | AvrA delL154 with IP6, CoA | | 分子名称: | AvrA, COENZYME A, INOSITOL HEXAKISPHOSPHATE | | 著者 | Labriola, J.M, Nagar, B. | | 登録日 | 2017-10-24 | | 公開日 | 2018-08-01 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.438 Å) | | 主引用文献 | Structural Analysis of the Bacterial Effector AvrA Identifies a Critical Helix Involved in Substrate Recognition.
Biochemistry, 57, 2018
|
|
6BVU
 
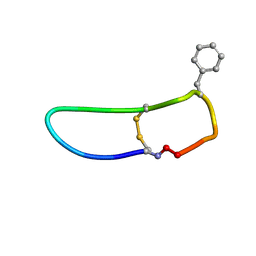 | | SFTI-HFRW-1 | | 分子名称: | Trypsin inhibitor 1 HFRW-1 | | 著者 | Schroeder, C.I. | | 登録日 | 2017-12-13 | | 公開日 | 2018-12-19 | | 最終更新日 | 2023-06-14 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Development of Novel Melanocortin Receptor Agonists Based on the Cyclic Peptide Framework of Sunflower Trypsin Inhibitor-1.
J.Med.Chem., 61, 2018
|
|
6LN1
 
 | | A natural inhibitor of DYRK1A for treatment of diabetes mellitus | | 分子名称: | 1,3,5,8-tetrakis(oxidanyl)xanthen-9-one, Dual specificity tyrosine-phosphorylation-regulated kinase 1A | | 著者 | Li, H, Chen, L.X, Zheng, M.Z, Zhang, Q.Z, Zhang, C.L, Wu, C.R, Yang, K.Y, Song, Z.R, Wang, Q.Q, Li, C, Zhou, Y.R, Chen, J.C. | | 登録日 | 2019-12-28 | | 公開日 | 2021-10-06 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.699 Å) | | 主引用文献 | A natural DYRK1A inhibitor as a potential stimulator for beta-cell proliferation in diabetes.
Clin Transl Med, 11, 2021
|
|
5Z1C
 
 | | The crystal structure of uPA in complex with 4-Iodobenzylamine at pH7.4 | | 分子名称: | 1-(4-iodophenyl)methanamine, Urokinase-type plasminogen activator | | 著者 | Jiang, L.G, Zhang, X, Luo, Z.P, Huang, M.D. | | 登録日 | 2017-12-25 | | 公開日 | 2018-12-26 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.45 Å) | | 主引用文献 | Halogen bonding for the design of inhibitors by targeting the S1 pocket of serine proteases
Rsc Adv, 49, 2018
|
|
6MT0
 
 | | Crystal structure of human Pim-1 kinase in complex with a quinazolinone-pyrrolodihydropyrrolone inhibitor | | 分子名称: | 3-(1-methylcyclopropyl)-2-[(1-methylcyclopropyl)amino]-8-[(6R)-6-methyl-4-oxo-1,4,5,6-tetrahydropyrrolo[3,4-b]pyrrol-2-yl]quinazolin-4(3H)-one, GLYCEROL, Serine/threonine-protein kinase pim-1 | | 著者 | Mohr, C. | | 登録日 | 2018-10-18 | | 公開日 | 2019-01-16 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Discovery of ( R)-8-(6-Methyl-4-oxo-1,4,5,6-tetrahydropyrrolo[3,4- b]pyrrol-2-yl)-3-(1-methylcyclopropyl)-2-((1-methylcyclopropyl)amino)quinazolin-4(3 H)-one, a Potent and Selective Pim-1/2 Kinase Inhibitor for Hematological Malignancies.
J. Med. Chem., 62, 2019
|
|
6BVW
 
 | | SFTI-HFRW-3 | | 分子名称: | Trypsin inhibitor 1 HFRW-3 | | 著者 | Schroeder, C.I. | | 登録日 | 2017-12-14 | | 公開日 | 2018-12-19 | | 最終更新日 | 2023-11-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Development of Novel Melanocortin Receptor Agonists Based on the Cyclic Peptide Framework of Sunflower Trypsin Inhibitor-1.
J.Med.Chem., 61, 2018
|
|
6BVX
 
 | | SFTI-HFRW-2 | | 分子名称: | Trypsin inhibitor 1 HFRW-2 | | 著者 | Schroeder, C.I. | | 登録日 | 2017-12-14 | | 公開日 | 2018-12-19 | | 最終更新日 | 2023-11-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Development of Novel Melanocortin Receptor Agonists Based on the Cyclic Peptide Framework of Sunflower Trypsin Inhibitor-1.
J.Med.Chem., 61, 2018
|
|
6BVY
 
 | | SFTI-HFRW-4 | | 分子名称: | Trypsin inhibitor 1 HFRW-4 | | 著者 | Schroeder, C.I, White, A. | | 登録日 | 2017-12-14 | | 公開日 | 2018-04-18 | | 最終更新日 | 2023-11-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Development of Novel Melanocortin Receptor Agonists Based on the Cyclic Peptide Framework of Sunflower Trypsin Inhibitor-1.
J. Med. Chem., 61, 2018
|
|
6K2Z
 
 | | Human Galectin-14 with lactose | | 分子名称: | Placental protein 13-like, beta-D-galactopyranose-(1-4)-alpha-D-glucopyranose, beta-D-galactopyranose-(1-4)-beta-D-glucopyranose | | 著者 | Su, J. | | 登録日 | 2019-05-15 | | 公開日 | 2020-06-17 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structure-function studies of galectin-14, an important effector molecule in embryology.
Febs J., 288, 2021
|
|
6KW1
 
 | | The structure of the metallo-beta-lactamase VIM-2 in complex with a triazolylthioacetamide 1b | | 分子名称: | 2-[3-[2-(1H-benzimidazol-2-ylamino)-2-oxidanylidene-ethyl]sulfanyl-1H-1,2,4-triazol-5-yl]benzoic acid, ACETATE ION, Beta-lactamase class B VIM-2, ... | | 著者 | Yang, K.W, Xiang, Y. | | 登録日 | 2019-09-05 | | 公開日 | 2020-09-09 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.775212 Å) | | 主引用文献 | Kinetic, Thermodynamic, and Crystallographic Studies of 2-Triazolylthioacetamides as Verona Integron-Encoded Metallo-beta-Lactamase 2 (VIM-2) Inhibitor.
Biomolecules, 10, 2020
|
|
6LJP
 
 | | Crystal structure of human galectin-16 | | 分子名称: | Galectin-16 | | 著者 | Su, J. | | 登録日 | 2019-12-17 | | 公開日 | 2020-10-14 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Human galectin-16 has a pseudo ligand binding site and plays a role in regulating c-Rel-mediated lymphocyte activity.
Biochim Biophys Acta Gen Subj, 1865, 2020
|
|
6LJQ
 
 | | human galectin-16 R55N | | 分子名称: | Galectin-16, beta-D-galactopyranose-(1-4)-beta-D-glucopyranose | | 著者 | Su, J. | | 登録日 | 2019-12-17 | | 公開日 | 2020-10-14 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.49 Å) | | 主引用文献 | Human galectin-16 has a pseudo ligand binding site and plays a role in regulating c-Rel-mediated lymphocyte activity.
Biochim Biophys Acta Gen Subj, 1865, 2020
|
|
6LJR
 
 | | human galectin-16 R55N/H57R | | 分子名称: | Galectin-16, beta-D-galactopyranose-(1-4)-beta-D-glucopyranose | | 著者 | Su, J. | | 登録日 | 2019-12-17 | | 公開日 | 2020-10-14 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Human galectin-16 has a pseudo ligand binding site and plays a role in regulating c-Rel-mediated lymphocyte activity.
Biochim Biophys Acta Gen Subj, 1865, 2020
|
|
6K2Y
 
 | | Human Galectin-14 | | 分子名称: | Placental protein 13-like | | 著者 | Su, J. | | 登録日 | 2019-05-15 | | 公開日 | 2020-06-17 | | 最終更新日 | 2021-03-10 | | 実験手法 | X-RAY DIFFRACTION (1.57 Å) | | 主引用文献 | Structure-function studies of galectin-14, an important effector molecule in embryology.
Febs J., 288, 2021
|
|
5EWS
 
 | | Sugar binding protein - human galectin-2 | | 分子名称: | Galectin-2, beta-D-galactopyranose-(1-4)-beta-D-glucopyranose | | 著者 | Su, J.Y, Si, Y.L. | | 登録日 | 2015-11-21 | | 公開日 | 2016-09-14 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Human galectin-2 interacts with carbohydrates and peptides non-classically: new insight from X-ray crystallography and hemagglutination.
Acta Biochim. Biophys. Sin. (Shanghai), 48, 2016
|
|
6OBD
 
 | |
6LBP
 
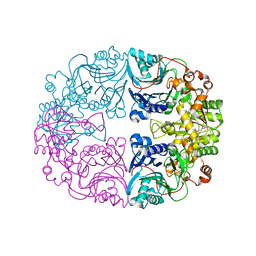 | | Structure of the Glutamine Phosphoribosylpyrophosphate Amidotransferase from Arabidopsis thaliana | | 分子名称: | Amidophosphoribosyltransferase 2, chloroplastic, IRON/SULFUR CLUSTER | | 著者 | Yi, Z, Cao, X, Han, F, Feng, Y. | | 登録日 | 2019-11-14 | | 公開日 | 2020-04-29 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (3.065 Å) | | 主引用文献 | Crystal Structure of the Chloroplastic Glutamine Phosphoribosylpyrophosphate Amidotransferase GPRAT2 FromArabidopsis thaliana.
Front Plant Sci, 11, 2020
|
|
4RPV
 
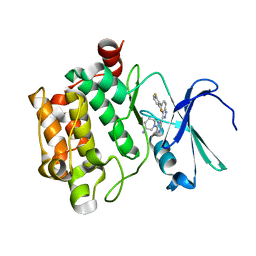 | | co-crystal structure of Pim1 with compound 3 | | 分子名称: | (3S)-1-{6-[5-(2,6-difluorophenyl)-2H-indazol-3-yl]pyrazin-2-yl}piperidin-3-amine, Serine/threonine-protein kinase pim-1 | | 著者 | Huang, X. | | 登録日 | 2014-10-31 | | 公開日 | 2015-02-04 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (3.05 Å) | | 主引用文献 | The discovery of novel 3-(pyrazin-2-yl)-1H-indazoles as potent pan-Pim kinase inhibitors.
Bioorg.Med.Chem.Lett., 25, 2015
|
|
5IPJ
 
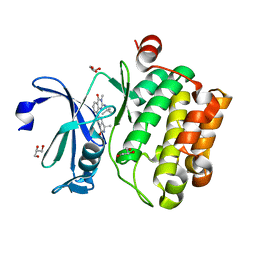 | |
5GQT
 
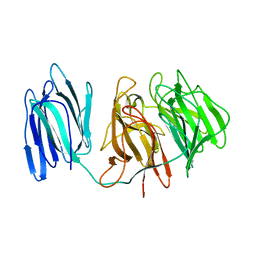 | |
5YLF
 
 | | MCR-1 complex with D-glucose | | 分子名称: | Probable phosphatidylethanolamine transferase Mcr-1, ZINC ION, beta-D-glucopyranose | | 著者 | Wei, P.C, Song, G.J, Shi, M.Y, Zhou, Y.F, Liu, Y, Lei, J, Chen, P, Yin, L. | | 登録日 | 2017-10-17 | | 公開日 | 2017-11-08 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Substrate analog interaction with MCR-1 offers insight into the rising threat of the plasmid-mediated transferable colistin resistance.
FASEB J., 32, 2018
|
|
5YLC
 
 | | Crystal Structure of MCR-1 Catalytic Domain | | 分子名称: | Probable phosphatidylethanolamine transferase Mcr-1, ZINC ION | | 著者 | Wei, P.C, Song, G.J, Shi, M.Y, Zhou, Y.F, Liu, Y, Lei, J, Chen, P, Yin, L. | | 登録日 | 2017-10-17 | | 公開日 | 2017-11-08 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Substrate analog interaction with MCR-1 offers insight into the rising threat of the plasmid-mediated transferable colistin resistance.
FASEB J., 32, 2018
|
|
