3IIV
 
 | |
3IIO
 
 | |
3IIP
 
 | |
5CY5
 
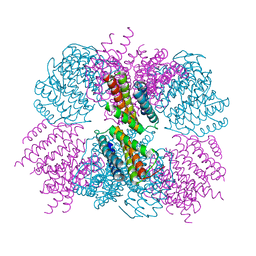 | | Crystal structure of the T33-51H designed self-assembling protein tetrahedron | | 分子名称: | T33-51H-A, T33-51H-B | | 著者 | Cannon, K.A, Cascio, D, Park, R, Boyken, S, King, N, Yeates, T.O. | | 登録日 | 2015-07-30 | | 公開日 | 2016-08-10 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (3.4 Å) | | 主引用文献 | Design and structure of two new protein cages illustrate successes and ongoing challenges in protein engineering.
Protein Sci., 29, 2020
|
|
3I1C
 
 | |
5IF6
 
 | |
7Q1V
 
 | |
7F1I
 
 | | Designed enzyme RA61 M48K/I72D mutant: form II | | 分子名称: | Engineered Retroaldolase | | 著者 | Fujioka, T, Oka, M, Numoto, N, Ito, N, Oda, M, Tanaka, F. | | 登録日 | 2021-06-09 | | 公開日 | 2021-11-24 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | Varying the Directionality of Protein Catalysts for Aldol and Retro-Aldol Reactions.
Chembiochem, 23, 2022
|
|
7F1H
 
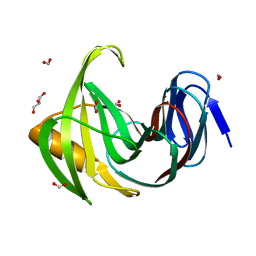 | | Designed enzyme RA61 M48K/I72D mutant: form I | | 分子名称: | Engineered Retroaldolase, FORMIC ACID, GLYCEROL | | 著者 | Fujioka, T, Oka, M, Numoto, N, Ito, N, Oda, M, Tanaka, F. | | 登録日 | 2021-06-09 | | 公開日 | 2021-11-24 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.14 Å) | | 主引用文献 | Varying the Directionality of Protein Catalysts for Aldol and Retro-Aldol Reactions.
Chembiochem, 23, 2022
|
|
7F1J
 
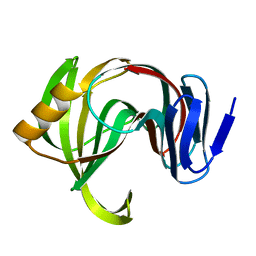 | | Designed enzyme RA61 M48K/I72D mutant: form III | | 分子名称: | Engineered Retroaldolase | | 著者 | Fujioka, T, Oka, M, Numoto, N, Ito, N, Oda, M, Tanaka, F. | | 登録日 | 2021-06-09 | | 公開日 | 2021-11-24 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Varying the Directionality of Protein Catalysts for Aldol and Retro-Aldol Reactions.
Chembiochem, 23, 2022
|
|
7F1K
 
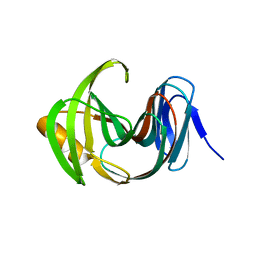 | | Designed enzyme RA61 M48K/I72D mutant: form IV | | 分子名称: | Engineered Retroaldolase | | 著者 | Fujioka, T, Oka, M, Numoto, N, Ito, N, Oda, M, Tanaka, F. | | 登録日 | 2021-06-09 | | 公開日 | 2021-11-24 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.05 Å) | | 主引用文献 | Varying the Directionality of Protein Catalysts for Aldol and Retro-Aldol Reactions.
Chembiochem, 23, 2022
|
|
7F1L
 
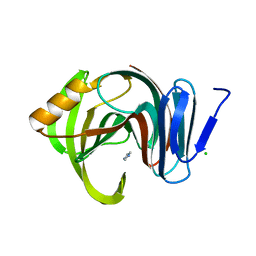 | | Designed enzyme RA61 M48K/I72D mutant: form V | | 分子名称: | CHLORIDE ION, Engineered Retroaldolase, IMIDAZOLE | | 著者 | Fujioka, T, Oka, M, Numoto, N, Ito, N, Oda, M, Tanaka, F. | | 登録日 | 2021-06-09 | | 公開日 | 2021-11-24 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Varying the Directionality of Protein Catalysts for Aldol and Retro-Aldol Reactions.
Chembiochem, 23, 2022
|
|
6VL5
 
 | |
6VL6
 
 | |
5JG9
 
 | |
5IER
 
 | |
6VKN
 
 | | BG505 SOSIP.v5.2.N241.N289 in complex with rhesus macaque Fab RM19R | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Cottrell, C.A, Ward, A.B. | | 登録日 | 2020-01-21 | | 公開日 | 2020-08-12 | | 最終更新日 | 2020-09-02 | | 実験手法 | ELECTRON MICROSCOPY (3.7 Å) | | 主引用文献 | Targeting HIV Env immunogens to B cell follicles in nonhuman primates through immune complex or protein nanoparticle formulations.
NPJ Vaccines, 5, 2020
|
|
5KWX
 
 | |
6WSJ
 
 | |
5IEP
 
 | |
5KWO
 
 | |
5KX2
 
 | |
5KWZ
 
 | |
6EGC
 
 | |
3NR6
 
 | | Crystal structure of xenotropic murine leukemia virus-related virus (XMRV) protease | | 分子名称: | PHOSPHATE ION, POTASSIUM ION, Protease p14 | | 著者 | Lubkowski, J, Li, M, Gustchina, A, Zhou, D, Dauter, Z, Wlodawer, A. | | 登録日 | 2010-06-30 | | 公開日 | 2011-02-02 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (1.97 Å) | | 主引用文献 | Crystal structure of XMRV protease differs from the structures of other retropepsins.
Nat.Struct.Mol.Biol., 18, 2011
|
|
