7RU6
 
 | |
7RUG
 
 | |
7RW4
 
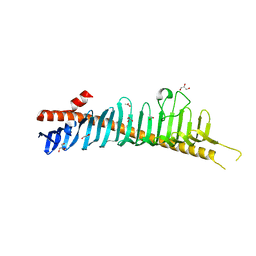 | | Crystal structure of junctophilin-1 | | 分子名称: | ACETATE ION, GLYCEROL, Junctophilin-1 | | 著者 | Yang, Z, Panwar, P, Van Petegem, F. | | 登録日 | 2021-08-19 | | 公開日 | 2022-02-23 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (1.31 Å) | | 主引用文献 | Structures of the junctophilin/voltage-gated calcium channel interface reveal hot spot for cardiomyopathy mutations.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7SPP
 
 | |
7SPO
 
 | |
7RXQ
 
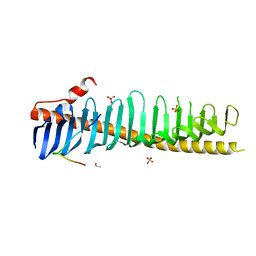 | | Crystal structure of junctophilin-2 in complex with a CaV1.1 peptide | | 分子名称: | ETHANOL, Junctophilin-2 N-terminal fragment, SULFATE ION, ... | | 著者 | Yang, Z, Panwar, P, Van Petegem, F. | | 登録日 | 2021-08-23 | | 公開日 | 2022-02-23 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.03 Å) | | 主引用文献 | Structures of the junctophilin/voltage-gated calcium channel interface reveal hot spot for cardiomyopathy mutations.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7RXE
 
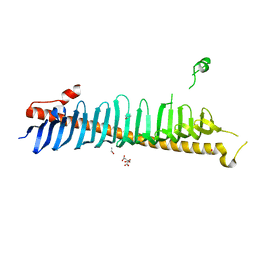 | | Crystal structure of junctophilin-2 | | 分子名称: | CITRATE ANION, ISOPROPYL ALCOHOL, Junctophilin-2 N-terminal fragment | | 著者 | Yang, Z, Panwar, P, Van Petegem, F. | | 登録日 | 2021-08-22 | | 公開日 | 2022-02-23 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.35 Å) | | 主引用文献 | Structures of the junctophilin/voltage-gated calcium channel interface reveal hot spot for cardiomyopathy mutations.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7SMU
 
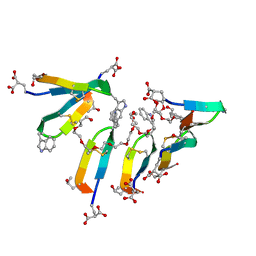 | | Crystal Structure of Consomatin-Ro1 | | 分子名称: | 3,6,9,12,15,18,21,24,27,30,33,36,39,42,45-pentadecaoxaoctatetracontane-1,48-diol, Consomatin-Ro1 | | 著者 | Ramiro, I.B.L, Whitby, F.G, Hill, C.P, Safavi-Hemami, H, Concepcion, G.P, Olivera, B.M. | | 登録日 | 2021-10-26 | | 公開日 | 2022-04-13 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Somatostatin venom analogs evolved by fish-hunting cone snails: From prey capture behavior to identifying drug leads.
Sci Adv, 8, 2022
|
|
7SQI
 
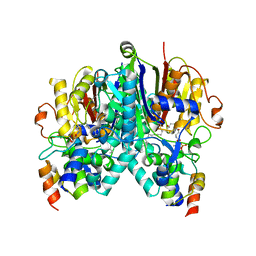 | | Crosslinked Crystal Structure of Type II Fatty Acid Synthase Ketosynthase, FabB, and C14-crypto Acyl Carrier Protein, AcpP | | 分子名称: | Acyl carrier protein, Beta-ketoacyl-ACP synthase I, N-{2-[(2Z)-3-chlorotetradec-2-enamido]ethyl}-N~3~-[(2R)-2-hydroxy-3,3-dimethyl-4-(phosphonooxy)butanoyl]-beta-alaninamide, ... | | 著者 | Chen, A, Mindrebo, J.T, Davis, T.D, Noel, J.P, Burkart, M.D. | | 登録日 | 2021-11-05 | | 公開日 | 2022-08-03 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Mechanism-based cross-linking probes capture the Escherichia coli ketosynthase FabB in conformationally distinct catalytic states.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7SZO
 
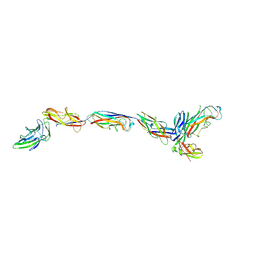 | | Structure of a bacterial fimbrial tip containing FocH | | 分子名称: | Chaperone protein FimC, FimF protein, FimG, ... | | 著者 | Stenkamp, R.E, Le Trong, I, Aprikian, P, Sokurenko, E.V. | | 登録日 | 2021-11-29 | | 公開日 | 2021-12-29 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Recombinant FimH Adhesin Demonstrates How the Allosteric Catch Bond Mechanism Can Support Fast and Strong Bacterial Attachment in the Absence of Shear.
J.Mol.Biol., 434, 2022
|
|
7SZ9
 
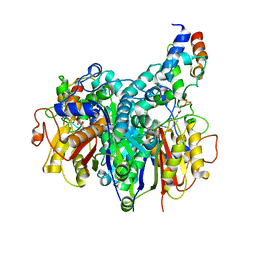 | | Crosslinked Crystal Structure of Type II Fatty Acid Synthase Ketosynthase, FabB, and C16:1-crypto Acyl Carrier Protein, AcpP | | 分子名称: | 3-oxoacyl-[acyl-carrier-protein] synthase 1, Acyl carrier protein, N~3~-{(2R)-4-[(dihydroxyphosphanyl)oxy]-2-hydroxy-3,3-dimethylbutanoyl}-N-(2-{[(9Z)-hexadec-9-enoyl]amino}ethyl)-beta-alaninamide, ... | | 著者 | Mindrebo, J.T, Chen, A, Noel, J.P, Burkart, M.D. | | 登録日 | 2021-11-26 | | 公開日 | 2022-11-23 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Mechanism-based cross-linking probes capture the Escherichia coli ketosynthase FabB in conformationally distinct catalytic states.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
1O68
 
 | |
1O6D
 
 | |
4G2L
 
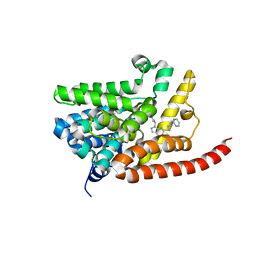 | | Human PDE9 in complex with selective compound | | 分子名称: | 1-cyclopentyl-6-{(1R)-1-[3-(pyrimidin-2-yl)azetidin-1-yl]ethyl}-1,5-dihydro-4H-pyrazolo[3,4-d]pyrimidin-4-one, High affinity cGMP-specific 3',5'-cyclic phosphodiesterase 9A, MAGNESIUM ION, ... | | 著者 | Liu, S. | | 登録日 | 2012-07-12 | | 公開日 | 2013-05-29 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Application of structure-based drug design and parallel chemistry to identify selective, brain penetrant, in vivo active phosphodiesterase 9A inhibitors.
J.Med.Chem., 55, 2012
|
|
4G2J
 
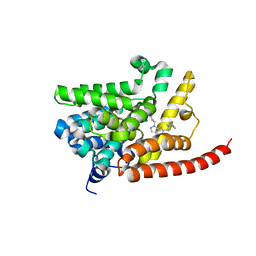 | | Human pde9 in complex with selective compound | | 分子名称: | 1-cyclopentyl-6-[(1R)-1-(3-phenoxyazetidin-1-yl)ethyl]-1,5-dihydro-4H-pyrazolo[3,4-d]pyrimidin-4-one, High affinity cGMP-specific 3',5'-cyclic phosphodiesterase 9A, MAGNESIUM ION, ... | | 著者 | Liu, S. | | 登録日 | 2012-07-12 | | 公開日 | 2013-05-29 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Application of structure-based drug design and parallel chemistry to identify selective, brain penetrant, in vivo active phosphodiesterase 9A inhibitors.
J.Med.Chem., 55, 2012
|
|
1O69
 
 | | Crystal structure of a PLP-dependent enzyme | | 分子名称: | (2-AMINO-4-FORMYL-5-HYDROXY-6-METHYLPYRIDIN-3-YL)METHYL DIHYDROGEN PHOSPHATE, BETA-MERCAPTOETHANOL, aminotransferase | | 著者 | Structural GenomiX | | 登録日 | 2003-10-23 | | 公開日 | 2003-11-11 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.84 Å) | | 主引用文献 | Structural analysis of a set of proteins resulting from a bacterial genomics project
Proteins, 60, 2005
|
|
1O62
 
 | |
1O64
 
 | |
1O65
 
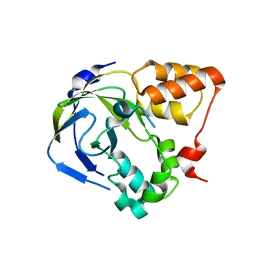 | |
1OQE
 
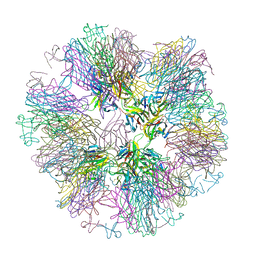 | | Crystal structure of sTALL-1 with BAFF-R | | 分子名称: | Tumor necrosis factor ligand superfamily member 13B, soluble form, Tumor necrosis factor receptor superfamily member 13C | | 著者 | Zhang, G. | | 登録日 | 2003-03-07 | | 公開日 | 2003-05-13 | | 最終更新日 | 2018-01-31 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Ligand-receptor binding revealed by the TNF family member TALL-1.
Nature, 423, 2003
|
|
1OQD
 
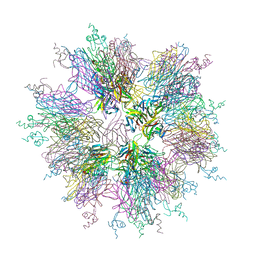 | | Crystal structure of sTALL-1 and BCMA | | 分子名称: | Tumor necrosis factor ligand superfamily member 13B, soluble form, Tumor necrosis factor receptor superfamily member 17 | | 著者 | Zhang, G. | | 登録日 | 2003-03-07 | | 公開日 | 2003-05-13 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Ligand-receptor binding revealed by the TNF family member TALL-1.
Nature, 423, 2003
|
|
4HTW
 
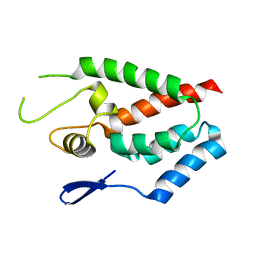 | | SIVmac239 capsid N-terminal domain | | 分子名称: | Gag protein | | 著者 | Schmidt, A.G. | | 登録日 | 2012-11-02 | | 公開日 | 2013-05-22 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Gain-of-Sensitivity Mutations in a Trim5-Resistant Primary Isolate of Pathogenic SIV Identify Two Independent Conserved Determinants of Trim5alpha Specificity.
Plos Pathog., 9, 2013
|
|
4L4Y
 
 | |
4L4Z
 
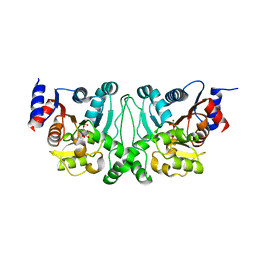 | | Crystal structures of the LsrR proteins complexed with phospho-AI-2 and its two different analogs reveal distinct mechanisms for ligand recognition | | 分子名称: | (2S)-2,3,3-trihydroxy-4-oxopentyl dihydrogen phosphate, Transcriptional regulator LsrR | | 著者 | Ryu, K.S, Ha, J.H, Eo, Y. | | 登録日 | 2013-06-10 | | 公開日 | 2013-11-06 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Crystal Structures of the LsrR Proteins Complexed with Phospho-AI-2 and Two Signal-Interrupting Analogues Reveal Distinct Mechanisms for Ligand Recognition.
J.Am.Chem.Soc., 135, 2013
|
|
4L50
 
 | | Crystal structures of the LsrR proteins complexed with phospho-AI-2 and its two different analogs reveal distinct mechanisms for ligand recognition | | 分子名称: | (2S)-2,3,3-trihydroxy-6-methyl-4-oxoheptyl dihydrogen phosphate, Transcriptional regulator LsrR | | 著者 | Ryu, K.S, Ha, J.H, Eo, Y. | | 登録日 | 2013-06-10 | | 公開日 | 2013-11-06 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Crystal Structures of the LsrR Proteins Complexed with Phospho-AI-2 and Two Signal-Interrupting Analogues Reveal Distinct Mechanisms for Ligand Recognition.
J.Am.Chem.Soc., 135, 2013
|
|
