1QIB
 
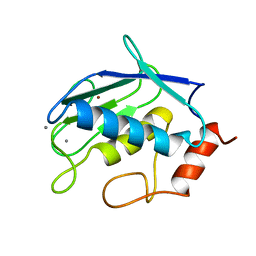 | | CRYSTAL STRUCTURE OF GELATINASE A CATALYTIC DOMAIN | | 分子名称: | 72 kDa type IV collagenase, CALCIUM ION, ZINC ION | | 著者 | Dhanaraj, V, Williams, M.G, Ye, Q.-Z, Molina, F, Johnson, L.L, Ortwine, D.F, Pavlovsky, A, Rubin, J.R, Skeean, R.W, White, A.D, Humblet, C, Hupe, D.J, Blundell, T.L. | | 登録日 | 1999-06-11 | | 公開日 | 1999-11-19 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | X-ray structure of gelatinase A catalytic domain complexed with a hydroxamate inhibitor
Croatica Chemica Acta, 72, 1999
|
|
4C0S
 
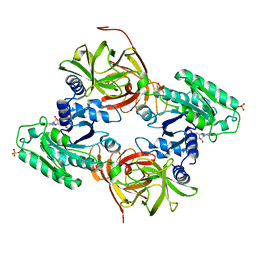 | | Mammalian translation elongation factor eEF1A2 | | 分子名称: | ELONGATION FACTOR 1-ALPHA 2, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION | | 著者 | Crepin, T, Shalak, V.F, Yaremchuk, A.D, Vlasenko, D.O, McCarthy, A.A, Negrutskii, B.S, Tukalo, M.A, El'skaya, A.V. | | 登録日 | 2013-08-07 | | 公開日 | 2014-08-27 | | 最終更新日 | 2017-01-25 | | 実験手法 | X-RAY DIFFRACTION (2.703 Å) | | 主引用文献 | Mammalian Translation Elongation Factor Eef1A2: X-Ray Structure and New Features of Gdp/GTP Exchange Mechanism in Higher Eukaryotes
Nucleic Acids Res., 42, 2014
|
|
4BWZ
 
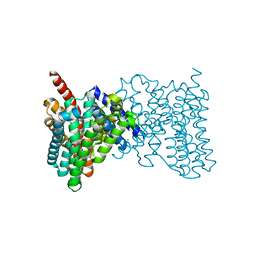 | |
1P8F
 
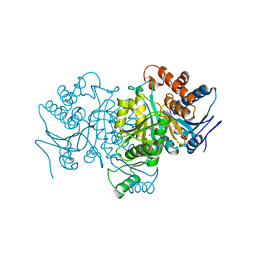 | |
1N2W
 
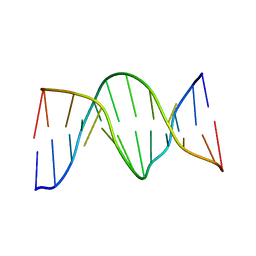 | | Solution Structure of 8OG:G mismatch containing duplex | | 分子名称: | 5'-D(*CP*GP*CP*GP*AP*AP*TP*TP*(8OG)P*GP*CP*G)-3' | | 著者 | Thiviyanathan, V, Somasunderam, A.D, Hazra, T.K, Mitra, S, Gorenstein, D.G. | | 登録日 | 2002-10-24 | | 公開日 | 2002-11-13 | | 最終更新日 | 2024-05-01 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution Structure of a DNA Duplex Containing 8-Hydroxy-2'-Deoxyguanosine Opposite Deoxyguanosine
J.Mol.Biol., 325, 2003
|
|
1QW7
 
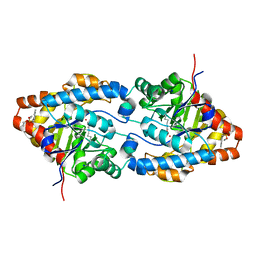 | | Structure of an Engineered Organophosphorous Hydrolase with Increased Activity Toward Hydrolysis of Phosphothiolate Bonds | | 分子名称: | COBALT (II) ION, DIETHYL 4-METHYLBENZYLPHOSPHONATE, Parathion hydrolase, ... | | 著者 | Mesecar, A.D, Grimsley, J.K, Holton, T, Wild, J.R. | | 登録日 | 2003-09-01 | | 公開日 | 2004-11-30 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Structural and mutational studies of organophosphorus hydrolase reveal a cryptic and functional allosteric-binding site.
Arch.Biochem.Biophys., 442, 2005
|
|
4ATV
 
 | | STRUCTURE OF A TRIPLE MUTANT OF THE NHAA DIMER, CRYSTALLISED AT LOW PH | | 分子名称: | DODECYL-ALPHA-D-MALTOSIDE, NA(+)/H(+) ANTIPORTER NHAA, SULFATE ION | | 著者 | Drew, D, Lee, C, Iwata, S, Cameron, A.D. | | 登録日 | 2012-05-10 | | 公開日 | 2013-07-10 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (3.5 Å) | | 主引用文献 | Crystal structure of the sodium-proton antiporter NhaA dimer and new mechanistic insights.
J. Gen. Physiol., 144, 2014
|
|
1QCW
 
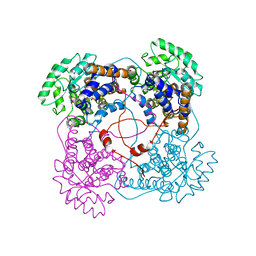 | | Flavocytochrome B2, ARG289LYS mutant | | 分子名称: | FLAVOCYTOCHROME B2, N-SULFO-FLAVIN MONONUCLEOTIDE | | 著者 | Mowat, C.G, Durley, R.C.E, Pike, A.D, Barton, J.D, Chen, Z.-W, Mathews, F.S, Lederer, F, Reid, G.A, Chapman, S.K. | | 登録日 | 1999-05-07 | | 公開日 | 1999-05-24 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2.75 Å) | | 主引用文献 | Kinetic and crystallographic studies on the active site Arg289Lys mutant of flavocytochrome b2 (yeast L-lactate dehydrogenase)
Biochemistry, 39, 2000
|
|
4B20
 
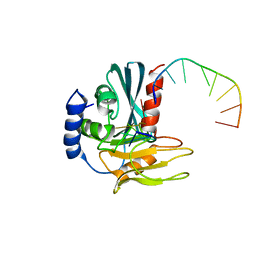 | | Structural basis of DNA loop recognition by Endonuclease V | | 分子名称: | 5'-D(*AP*TP*CP*TP*TP*GP*TP*CP*GP*CP)-3', 5'-D(*GP*CP*GP*AP*CP*AP*GP)-3', ENDONUCLEASE V, ... | | 著者 | Rosnes, I, Rowe, A.D, Forstrom, R.J, Alseth, I, Bjoras, M, Dalhus, B. | | 登録日 | 2012-07-12 | | 公開日 | 2013-04-17 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.75 Å) | | 主引用文献 | Structural Basis of DNA Loop Recognition by Endonuclease V.
Structure, 21, 2013
|
|
1QH5
 
 | | HUMAN GLYOXALASE II WITH S-(N-HYDROXY-N-BROMOPHENYLCARBAMOYL)GLUTATHIONE | | 分子名称: | GLUTATHIONE, PROTEIN (HYDROXYACYLGLUTATHIONE HYDROLASE), S-(N-HYDROXY-N-BROMOPHENYLCARBAMOYL)GLUTATHIONE, ... | | 著者 | Cameron, A.D, Ridderstrom, M, Olin, B, Mannervik, B. | | 登録日 | 1999-05-11 | | 公開日 | 1999-09-24 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.45 Å) | | 主引用文献 | Crystal structure of human glyoxalase II and its complex with a glutathione thiolester substrate analogue.
Structure Fold.Des., 7, 1999
|
|
1QIC
 
 | | CRYSTAL STRUCTURE OF STROMELYSIN CATALYTIC DOMAIN | | 分子名称: | CALCIUM ION, PROTEIN (STROMELYSIN-1), ZINC ION | | 著者 | Williams, M.G, Ye, Q.-Z, Molina, F, Johnson, L.L, Ortwine, D.F, Pavlovsky, A.G, Rubin, J.R, Skeean, R.W, White, A.D, Blundell, T.L, Humblet, C, Hupe, D.J, Dhanaraj, V. | | 登録日 | 1999-06-11 | | 公開日 | 2003-02-11 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | X-ray structure of human stromelysin catalytic domain complexed with nonpeptide inhibitors: implications for inhibitor selectivity
Protein Sci., 8, 1999
|
|
4D1D
 
 | | STRUCTURE OF MHP1, A NUCLEOBASE-CATION-SYMPORT-1 FAMILY TRANSPORTER with the inhibitor 5-(2-naphthylmethyl)-L-hydantoin. | | 分子名称: | 5-(2-NAPHTHYLMETHYL)-D-HYDANTOIN, 5-(2-NAPHTHYLMETHYL)-L-HYDANTOIN, HYDANTOIN TRANSPORT PROTEIN, ... | | 著者 | Weyand, S, Brueckner, F, Geng, T, Drew, D, Iwata, S, Henderson, P.J.F, Cameron, A.D. | | 登録日 | 2014-05-01 | | 公開日 | 2014-07-02 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (3.7 Å) | | 主引用文献 | Molecular Mechanism of Ligand Recognition by Membrane Transport Protein, Mhp1.
Embo J., 33, 2014
|
|
4D1A
 
 | | STRUCTURE OF MHP1, A NUCLEOBASE-CATION-SYMPORT-1 FAMILY TRANSPORTER, IN A CLOSED CONFORMATION WITH INDOLYLMETHYL-HYDANTOIN | | 分子名称: | (5S)-5-(1H-indol-3-ylmethyl)imidazolidine-2,4-dione, HYDANTOIN TRANSPORT PROTEIN, SODIUM ION | | 著者 | Weyand, S, Brueckner, F, Geng, T, Drew, D, Iwata, S, Henderson, P.J.F, Cameron, A.D. | | 登録日 | 2014-05-01 | | 公開日 | 2014-07-02 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (3.4 Å) | | 主引用文献 | Molecular Mechanism of Ligand Recognition by Membrane Transport Protein, Mhp1.
Embo J., 33, 2014
|
|
4D1B
 
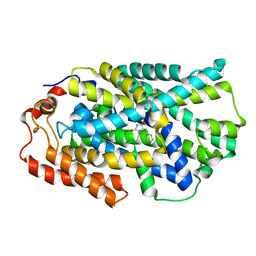 | | STRUCTURE OF MHP1, A NUCLEOBASE-CATION-SYMPORT-1 FAMILY TRANSPORTER, IN A CLOSED CONFORMATION WITH BENZYL-HYDANTOIN | | 分子名称: | (5S)-5-benzylimidazolidine-2,4-dione, HYDANTOIN TRANSPORT PROTEIN, SODIUM ION | | 著者 | Brueckner, F, Geng, T, Weyand, S, Drew, D, Iwata, S, Henderson, P.J.F, Cameron, A.D. | | 登録日 | 2014-05-01 | | 公開日 | 2014-07-02 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (3.8 Å) | | 主引用文献 | Molecular Mechanism of Ligand Recognition by Membrane Transport Protein, Mhp1.
Embo J., 33, 2014
|
|
1QIA
 
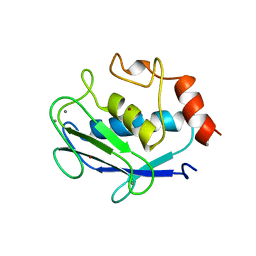 | | CRYSTAL STRUCTURE OF STROMELYSIN CATALYTIC DOMAIN | | 分子名称: | CALCIUM ION, STROMELYSIN-1, ZINC ION | | 著者 | Williams, M.G, Ye, Q.-Z, Molina, F, Johnson, L.L, Ortwine, D.F, Pavlovsky, A.G, Rubin, J.R, Skeean, R.W, White, A.D, Blundell, T.L, Humblet, C, Hupe, D.J, Dhanaraj, V. | | 登録日 | 1999-06-11 | | 公開日 | 2003-02-11 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | X-ray structure of human stromelysin catalytic domain complexed with nonpeptide inhibitors: implications for inhibitor selectivity
Protein Sci., 8, 1999
|
|
4EDJ
 
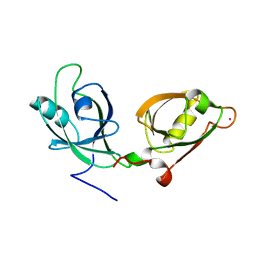 | | Crystal structure of the GRASP55 GRASP Domain with a phosphomimetic mutation (S189D) | | 分子名称: | Golgi reassembly-stacking protein 2, POTASSIUM ION | | 著者 | Truschel, S.T, Zhang, M, Bachert, C, Macbeth, M.R, Linstedt, A.D. | | 登録日 | 2012-03-27 | | 公開日 | 2012-05-09 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.901 Å) | | 主引用文献 | Allosteric Regulation of GRASP Protein-dependent Golgi Membrane Tethering by Mitotic Phosphorylation.
J.Biol.Chem., 287, 2012
|
|
4ERJ
 
 | |
1QH3
 
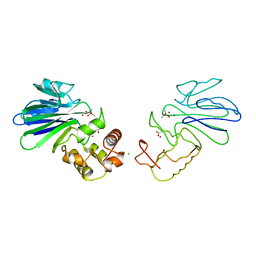 | | HUMAN GLYOXALASE II WITH CACODYLATE AND ACETATE IONS PRESENT IN THE ACTIVE SITE | | 分子名称: | ACETATE ION, CACODYLATE ION, CHLORIDE ION, ... | | 著者 | Cameron, A.D, Ridderstrom, M, Olin, B, Mannervik, B. | | 登録日 | 1999-05-10 | | 公開日 | 1999-09-24 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Crystal structure of human glyoxalase II and its complex with a glutathione thiolester substrate analogue.
Structure Fold.Des., 7, 1999
|
|
2J94
 
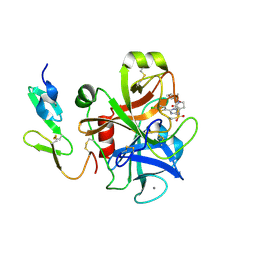 | | CRYSTAL STRUCTURE OF A HUMAN FACTOR XA INHIBITOR COMPLEX | | 分子名称: | 5-(5-CHLORO-2-THIENYL)-N-{(3S)-1-[(1S)-1-METHYL-2-MORPHOLIN-4-YL-2-OXOETHYL]-2-OXOPYRROLIDIN-3-YL}-1H-1,2,4-TRIAZOLE-3-SULFONAMIDE, CALCIUM ION, COAGULATION FACTOR X | | 著者 | Chan, C, Borthwick, A.D, Brown, D, Campbell, M, Chaudry, L, Chung, C.W, Convery, M.A, Hamblin, J.N, Johnstone, L, Kelly, H.A, Kleanthous, S, Burns-Kurtis, C.L, Patikis, A, Patel, C, Pateman, A.J, Senger, S, Shah, G.P, Toomey, J.R, Watson, N.S, Weston, H.E, Whitworth, C, Young, R.J, Zhou, P. | | 登録日 | 2006-11-02 | | 公開日 | 2007-03-20 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Factor Xa Inhibitors: S1 Binding Interactions of a Series of N-{(3S)-1-[(1S)-1-Methyl-2-Morpholin-4-Yl-2-Oxoethyl]-2-Oxopyrrolidin-3-Yl}Sulfonamides.
J.Med.Chem., 50, 2007
|
|
4GGJ
 
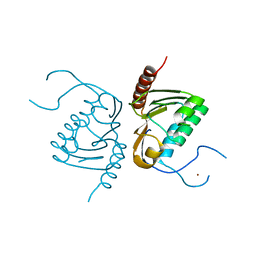 | | Crystal structure of Zucchini from mouse (mZuc / PLD6 / MitoPLD) | | 分子名称: | Mitochondrial cardiolipin hydrolase, ZINC ION | | 著者 | Ipsaro, J.J, Haase, A.D, Hannon, G.J, Joshua-Tor, L. | | 登録日 | 2012-08-06 | | 公開日 | 2012-10-10 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | The structural biochemistry of Zucchini implicates it as a nuclease in piRNA biogenesis.
Nature, 491, 2012
|
|
2J50
 
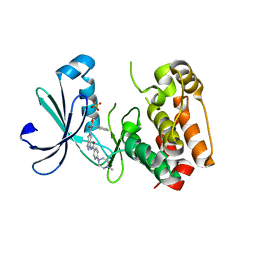 | | Structure of Aurora-2 in complex with PHA-739358 | | 分子名称: | N-[(3E)-5-[(2R)-2-METHOXY-2-PHENYLACETYL]PYRROLO[3,4-C]PYRAZOL-3(5H)-YLIDENE]-4-(4-METHYLPIPERAZIN-1-YL)BENZAMIDE, SERINE/THREONINE-PROTEIN KINASE 6, SULFATE ION | | 著者 | Cameron, A.D, Izzo, G, Storici, P, Rusconi, L, Fancelli, D, Varasi, M, Berta, D, Bindi, S, Forte, B, Severino, D, Tonani, R, Vianello, P. | | 登録日 | 2006-09-08 | | 公開日 | 2006-11-06 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | 1,4,5,6-Tetrahydropyrrolo[3,4-C]Pyrazoles: Identification of a Potent Aurora Kinase Inhibitor with a Favorable Antitumor Kinase Inhibition Profile.
J.Med.Chem., 49, 2006
|
|
2J4I
 
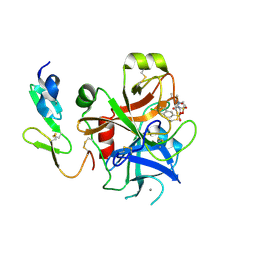 | | CRYSTAL STRUCTURE OF A HUMAN FACTOR XA INHIBITOR COMPLEX | | 分子名称: | 1-PYRROLIDINEACETAMIDE, 3-[[(6-CHLORO-2-NAPHTHALENYL)SULFONYL]AMINO]-ALPHA-METHYL-N-(1-METHYLETHYL)-N-[2-[(METHYLSULFONYL)AMINO]ETHYL]-2-OXO-, (ALPHAS,3S)-, ... | | 著者 | Young, R.J, Campbell, M, Borthwick, A.D, Brown, D, Chan, C, Convery, M.A, Crowe, M.C, Dayal, S, Diallo, H, Kelly, H.A, Paul King, N, Kleanthous, S, Kurtis, C.L, Mason, A.M, Mordaunt, J.E, Patel, C, Pateman, A.J, Senger, S, Shah, G.P, Smith, P.W, Watson, N.S, Weston, H.E, Zhou, P. | | 登録日 | 2006-08-31 | | 公開日 | 2006-09-27 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Structure- and Property-Based Design of Factor Xa Inhibitors: Pyrrolidin-2-Ones with Acyclic Alanyl Amides as P4 Motifs.
Bioorg.Med.Chem.Lett., 16, 2006
|
|
4GAV
 
 | | Structure of the Ndi1 protein from Saccharomyces cerevisiae in complex with quinone | | 分子名称: | FLAVIN-ADENINE DINUCLEOTIDE, Rotenone-insensitive NADH-ubiquinone oxidoreductase, UBIQUINONE-2 | | 著者 | Iwata, M, Lee, Y, Yamashita, T, Yagi, T, Iwata, S, Cameron, A.D, Maher, M.J. | | 登録日 | 2012-07-25 | | 公開日 | 2012-09-05 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | The structure of the yeast NADH dehydrogenase (Ndi1) reveals overlapping binding sites for water- and lipid-soluble substrates.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4G9K
 
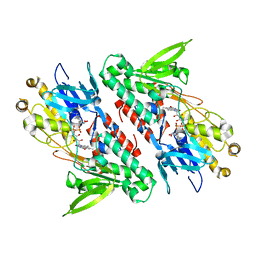 | | Structure of the Ndi1 protein from Saccharomyces cerevisiae | | 分子名称: | FLAVIN-ADENINE DINUCLEOTIDE, Rotenone-insensitive NADH-ubiquinone oxidoreductase | | 著者 | Iwata, M, Lee, Y, Yamashita, T, Yagi, T, Iwata, S, Cameron, A.D, Maher, M.J. | | 登録日 | 2012-07-24 | | 公開日 | 2012-09-05 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | The structure of the yeast NADH dehydrogenase (Ndi1) reveals overlapping binding sites for water- and lipid-soluble substrates.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4GAP
 
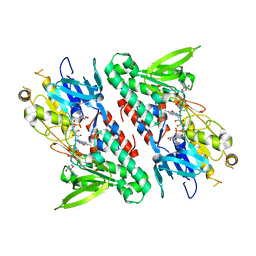 | | Structure of the Ndi1 protein from Saccharomyces cerevisiae in complex with NAD+ | | 分子名称: | FLAVIN-ADENINE DINUCLEOTIDE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, Rotenone-insensitive NADH-ubiquinone oxidoreductase | | 著者 | Iwata, M, Lee, Y, Yamashita, T, Yagi, T, Iwata, S, Cameron, A.D, Maher, M.J. | | 登録日 | 2012-07-25 | | 公開日 | 2012-09-05 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | The structure of the yeast NADH dehydrogenase (Ndi1) reveals overlapping binding sites for water- and lipid-soluble substrates.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
