4ZOQ
 
 | |
8CWX
 
 | |
8CZK
 
 | |
8CZL
 
 | |
8D19
 
 | | Human LanCL1 bound to GSH | | 分子名称: | GLUTATHIONE, Glutathione S-transferase LANCL1, ZINC ION | | 著者 | Ongpipattanakul, C, Nair, S.K. | | 登録日 | 2022-05-26 | | 公開日 | 2023-01-25 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.52 Å) | | 主引用文献 | The mechanism of thia-Michael addition catalyzed by LanC enzymes.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
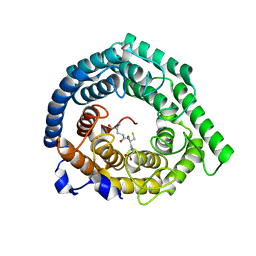
8D0V
 
 | |
6UAK
 
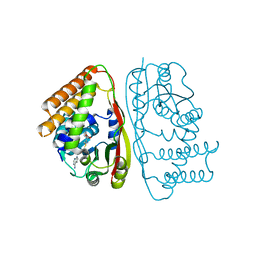 | |
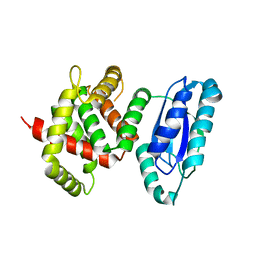
5WA3
 
 | |
5W99
 
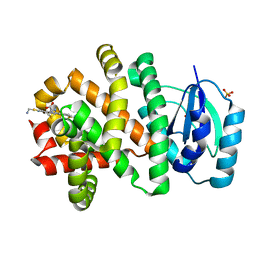 | | Pyridine synthase, PbtD, from GE2270 biosynthesis bound to TSP | | 分子名称: | 2,2'-(6-(2'-(aminomethyl)-[2,4'-bithiazol]-4-yl)pyridine-2,5-diyl)bis(thiazole-4-carboxylic acid), PbtD, SULFATE ION | | 著者 | Cogan, D.P, Nair, S.K. | | 登録日 | 2017-06-22 | | 公開日 | 2017-11-22 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.59 Å) | | 主引用文献 | Structural insights into enzymatic [4+2] aza-cycloaddition in thiopeptide antibiotic biosynthesis.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5WA4
 
 | |
5W98
 
 | |
6B9T
 
 | |
6B9S
 
 | |
6B9R
 
 | |
6C0Y
 
 | |
6C0G
 
 | |
6C0H
 
 | |
6EC7
 
 | |
6EC8
 
 | |
5DZT
 
 | |
5EHK
 
 | |
4QEC
 
 | | ElxO with NADP Bound | | 分子名称: | ElxO, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, SULFATE ION | | 著者 | Garg, N, Nair, S.K. | | 登録日 | 2014-05-15 | | 公開日 | 2014-07-02 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Substrate Specificity of the Lanthipeptide Peptidase ElxP and the Oxidoreductase ElxO.
Acs Chem.Biol., 9, 2014
|
|
4QED
 
 | | ElxO Y152F with NADPH Bound | | 分子名称: | ElxO, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, SULFATE ION | | 著者 | Garg, N, Nair, S.K. | | 登録日 | 2014-05-15 | | 公開日 | 2014-07-02 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Substrate Specificity of the Lanthipeptide Peptidase ElxP and the Oxidoreductase ElxO.
Acs Chem.Biol., 9, 2014
|
|
3RZZ
 
 | |
3SZZ
 
 | |
